Detecting adaptive convergent amino acid evolution
- PMID: 31154974
- PMCID: PMC6560273
- DOI: 10.1098/rstb.2018.0234
Detecting adaptive convergent amino acid evolution
Abstract
In evolutionary genomics, researchers have taken an interest in identifying substitutions that subtend convergent phenotypic adaptations. This is a difficult question that requires distinguishing foreground convergent substitutions that are involved in the convergent phenotype from background convergent substitutions. Those may be linked to other adaptations, may be neutral or may be the consequence of mutational biases. Furthermore, there is no generally accepted definition of convergent substitutions. Various methods that use different definitions have been proposed in the literature, resulting in different sets of candidate foreground convergent substitutions. In this article, we first describe the processes that can generate foreground convergent substitutions in coding sequences, separating adaptive from non-adaptive processes. Second, we review methods that have been proposed to detect foreground convergent substitutions in coding sequences and expose the assumptions that underlie them. Finally, we examine their power on simulations of convergent changes-including in the presence of a change in the efficacy of selection-and on empirical alignments. This article is part of the theme issue 'Convergent evolution in the genomics era: new insights and directions'.
Keywords: C3/C4; convergent evolution; genomics; molecular evolution; phylogenetics; probabilistic models.
Conflict of interest statement
We have no competing interests.
Figures
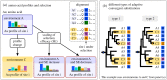
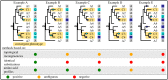
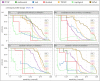


References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
