Comprehensive Modeling of Spinal Muscular Atrophy in Drosophila melanogaster
- PMID: 31156382
- PMCID: PMC6532329
- DOI: 10.3389/fnmol.2019.00113
Comprehensive Modeling of Spinal Muscular Atrophy in Drosophila melanogaster
Abstract
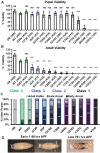
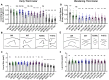
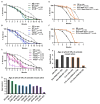
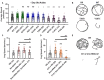
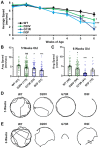
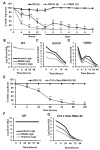
Spinal muscular atrophy (SMA) is a neurodegenerative disorder that affects motor neurons, primarily in young children. SMA is caused by mutations in the Survival Motor Neuron 1 (SMN1) gene. SMN functions in the assembly of spliceosomal RNPs and is well conserved in many model systems including mouse, zebrafish, fruit fly, nematode, and fission yeast. Work in Drosophila has focused on the loss of SMN function during larval stages, primarily using null alleles or strong hypomorphs. A systematic analysis of SMA-related phenotypes in the context of moderate alleles that more closely mimic the genetics of SMA has not been performed in the fly, leading to debate over the validity and translational value of this model. We, therefore, examined 14 Drosophila lines expressing SMA patient-derived missense mutations in Smn, with a focus on neuromuscular phenotypes in the adult stage. Animals were evaluated on the basis of organismal viability and longevity, locomotor function, neuromuscular junction structure, and muscle health. In all cases, we observed phenotypes similar to those of SMA patients, including progressive loss of adult motor function. The severity of these defects is variable and forms a broad spectrum across the 14 lines examined, recapitulating the full range of phenotypic severity observed in human SMA. This includes late-onset models of SMA, which have been difficult to produce in other model systems. The results provide direct evidence that SMA-related locomotor decline can be reproduced in the fly and support the use of patient-derived SMN missense mutations as a comprehensive system for modeling SMA.
Keywords: SMA; SMN1; SMN2; invertebrate models; neuromuscular disease; spinal muscular atrophy.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

