Salmonella Establishment in Agricultural Soil and Colonization of Crop Plants Depend on Soil Type and Plant Species
- PMID: 31156568
- PMCID: PMC6529577
- DOI: 10.3389/fmicb.2019.00967
Salmonella Establishment in Agricultural Soil and Colonization of Crop Plants Depend on Soil Type and Plant Species
Abstract
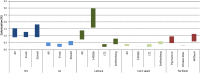
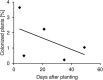
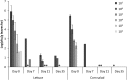
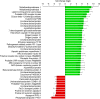
Human pathogenic bacteria, such as Salmonella enterica, are able to colonize crop plants. So far, not much is known about biotic and abiotic factors influencing this colonization in field soil. This understanding, however, is imperative for the provision of safe fresh produce to the consumer. In this study, we investigated the effects of soil type, organic fertilization, plant species and the way of Salmonella entry into the plant production system, on the survival of S. enterica in soil as well as the colonization of plants. The selected S. enterica serovar Typhimurium strain 14028s, S. Typhimurium strain LT2 and S. Senftenberg were able to persist in soil for several weeks. Salmonella's persistence in soil was prolonged in loamy, if compared to sandy soil, and when applied together with organic fertilizer. The leaves of lettuce and corn salad were colonized by S. enterica providing evidence for internalization from the soil via the root. Colonization rates were affected by soil type, plant species and S. enterica strain. Overall, S. enterica was detected in leaves of 0.5-0.9% of the plants, while lettuce was more frequently colonized than corn salad. Plants grown in sandy soil were more often colonized than plants grown in loamy soil. After spray inoculation, S. enterica could be detected on and in leaves for several weeks by cultivation-depending methods, confirmed by confocal microscopy using GFP-labeled S. Typhimurium 14028s. On the one hand, transcriptome data from S. Typhimurium 14028s assessed in response to lettuce medium or lettuce root exudates showed an upregulation of genes associated with biofilm formation and virulence. On the other hand, lettuce inoculated with S. Typhimurium 14028s showed a strong upregulation of genes associated with plant immune response and genes related to stress response. In summary, these results showed that organic fertilizers can increase the persistence of Salmonella in soil and that soil type and plant species play a crucial role in the interactions between human pathogens and crop plants. This understanding is therefore a starting point for new strategies to provide safe food for the consumer.
Keywords: Salmonella; crop plants; internalization; persistence; plant defense; soil.
Figures




References
-
- Basak B. B., Pal S., Datta S. C. (2012). Use of modified clays for retention and supply of water and nutrients. Curr. Sci. 102 1272–1278.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

