Sweetpotato bZIP Transcription Factor IbABF4 Confers Tolerance to Multiple Abiotic Stresses
- PMID: 31156685
- PMCID: PMC6531819
- DOI: 10.3389/fpls.2019.00630
Sweetpotato bZIP Transcription Factor IbABF4 Confers Tolerance to Multiple Abiotic Stresses
Abstract
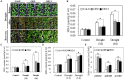
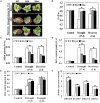
The abscisic acid (ABA)-responsive element binding factors (ABFs) play important regulatory roles in multiple abiotic stresses responses. However, information on the stress tolerance functions of ABF genes in sweetpotato (Ipomoea batatas [L.] Lam) remains limited. In the present study, we isolated and functionally characterized the sweetpotato IbABF4 gene, which encodes an abiotic stress-inducible basic leucine zipper (bZIP) transcription factor. Sequence analysis showed that the IbABF4 protein contains a typical bZIP domain and five conserved Ser/Thr kinase phosphorylation sites (RXXS/T). The IbABF4 gene was constitutively expressed in leaf, petiole, stem, and root, with the highest expression in storage root body. Expression of IbABF4 was induced by ABA and several environmental stresses including drought, salt, and heat shock. The IbABF4 protein localized to the nucleus, exhibited transcriptional activation activity, and showed binding to the cis-acting ABA-responsive element (ABRE) in vitro. Overexpression of IbABF4 in Arabidopsis thaliana not only increased ABA sensitivity but also enhanced drought and salt stress tolerance. Furthermore, transgenic sweetpotato plants (hereafter referred to as SA plants) overexpressing IbABF4, generated in this study, exhibited increased tolerance to drought, salt, and oxidative stresses on the whole plant level. This phenotype was associated with higher photosynthetic efficiency and lower malondialdehyde and hydrogen peroxide content. Levels of endogenous ABA content and ABA/stress-responsive gene expression were significantly upregulated in transgenic Arabidopsis and sweetpotato plants compared with wild-type plants under drought stress. Our results suggest that the expression of IbABF4 in Arabidopsis and sweetpotato enhances tolerance to multiple abiotic stresses through the ABA signaling pathway.
Keywords: IbABF4; abiotic stress; drought tolerance; salt tolerance; sweetpotato.
Figures









References
-
- Agarwal P. K., Jha B. (2010). Transcription factors in plants and ABA dependent and independent abiotic stress signaling. Biol. Plant 54 201–212. 10.1007/s10535-010-0038-7 - DOI
-
- Bartels D., Sunkar R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24 23–58. 10.1080/07352680590910410 - DOI
LinkOut - more resources
Full Text Sources

