RNA Next-Generation Sequencing and a Bioinformatics Pipeline to Identify Expressed LINE-1s at the Locus-Specific Level
- PMID: 31157783
- PMCID: PMC7371004
- DOI: 10.3791/59771
RNA Next-Generation Sequencing and a Bioinformatics Pipeline to Identify Expressed LINE-1s at the Locus-Specific Level
Abstract
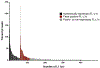
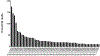
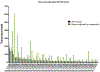
Long INterspersed Elements-1 (LINEs/L1s) are repetitive elements that can copy and randomly insert in the genome resulting in genomic instability and mutagenesis. Understanding the expression patterns of L1 loci at the individual level will lend to the understanding of the biology of this mutagenic element. This autonomous element makes up a significant portion of the human genome with over 500,000 copies, though 99% are truncated and defective. However, their abundance and dominant number of defective copies make it challenging to identify authentically expressed L1s from L1-related sequences expressed as part of other genes. It is also challenging to identify which specific L1 locus is expressed due to the repetitive nature of the elements. Overcoming these challenges, we present an RNA-Seq bioinformatic approach to identify L1 expression at the locus specific level. In summary, we collect cytoplasmic RNA, select for polyadenylated transcripts, and utilize strand-specific RNA-Seq analyses to uniquely map reads to L1 loci in the human reference genome. We visually curate each L1 locus with uniquely mapped reads to confirm transcription from its own promoter and adjust mapped transcript reads to account for mappability of each individual L1 locus. This approach was applied to a prostate tumor cell line, DU145, to demonstrate the ability of this protocol to detect expression from a small number of the full-length L1 elements.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
