Analysis of hereditary cancer syndromes by using a panel of genes: novel and multiple pathogenic mutations
- PMID: 31159747
- PMCID: PMC6547505
- DOI: 10.1186/s12885-019-5756-4
Analysis of hereditary cancer syndromes by using a panel of genes: novel and multiple pathogenic mutations
Abstract
Background: Hereditary cancer predisposition syndromes are responsible for approximately 5-10% of all diagnosed cancer cases. In the past, single-gene analysis of specific high risk genes was used for the determination of the genetic cause of cancer heritability in certain families. The application of Next Generation Sequencing (NGS) technology has facilitated multigene panel analysis and is widely used in clinical practice, for the identification of individuals with cancer predisposing gene variants. The purpose of this study was to investigate the extent and nature of variants in genes implicated in hereditary cancer predisposition in individuals referred for testing in our laboratory.
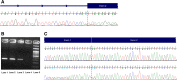
Methods: In total, 1197 individuals from Greece, Romania and Turkey were referred to our laboratory for genetic testing in the past 4 years. The majority of referrals included individuals with personal of family history of breast and/or ovarian cancer. The analysis of genes involved in hereditary cancer predisposition was performed using a NGS approach. Genomic DNA was enriched for targeted regions of 36 genes and sequencing was carried out using the Illumina NGS technology. The presence of large genomic rearrangements (LGRs) was investigated by computational analysis and Multiplex Ligation-dependent Probe Amplification (MLPA).
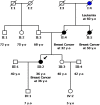
Results: A pathogenic variant was identified in 264 of 1197 individuals (22.1%) analyzed while a variant of uncertain significance (VUS) was identified in 34.8% of cases. Clinically significant variants were identified in 29 of the 36 genes analyzed. Concerning the mutation distribution among individuals with positive findings, 43.6% were located in the BRCA1/2 genes whereas 21.6, 19.9, and 15.0% in other high, moderate and low risk genes respectively. Notably, 25 of the 264 positive individuals (9.5%) carried clinically significant variants in two different genes and 6.1% had a LGR.
Conclusions: In our cohort, analysis of all the genes in the panel allowed the identification of 4.3 and 8.1% additional pathogenic variants in other high or moderate/low risk genes, respectively, enabling personalized management decisions for these individuals and supporting the clinical significance of multigene panel analysis in hereditary cancer predisposition.
Keywords: BRCA1 & BRCA2; Breast cancer; Cancer susceptibility genes; Genetic testing; Hereditary cancer; Large genomic rearrangement; Multigene panels; Next generation sequencing; Pathogenic variant.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Nagy R, Sweet K, Eng C. Highly penetrant hereditary cancer syndromes. Oncogene. 2004;23(38):6445–6470. - PubMed
-
- Garber JE, Offit K. Hereditary cancer predisposition syndromes. J Clin Oncol. 2005;23(2):276–292. - PubMed
-
- Susswein LR, Marshall ML, Nusbaum R, Vogel Postula KJ, Weissman SM, Yackowski L, Vaccari EM, Bissonnette J, Booker JK, Cremona ML, et al. Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing. Genet Med. 2016;18(8):823–832. - PMC - PubMed
-
- Azzariti Danielle R., Riggs Erin Rooney, Niehaus Annie, Rodriguez Laura Lyman, Ramos Erin M., Kattman Brandi, Landrum Melissa J., Martin Christa L., Rehm Heidi L. Points to consider for sharing variant-level information from clinical genetic testing with ClinVar. Molecular Case Studies. 2018;4(1):a002345. - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

