Automated extraction of sudden cardiac death risk factors in hypertrophic cardiomyopathy patients by natural language processing
- PMID: 31160009
- PMCID: PMC6550341
- DOI: 10.1016/j.ijmedinf.2019.05.008
Automated extraction of sudden cardiac death risk factors in hypertrophic cardiomyopathy patients by natural language processing
Abstract
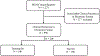
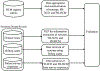
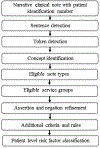
Background: The management of hypertrophic cardiomyopathy (HCM) patients requires the knowledge of risk factors associated with sudden cardiac death (SCD). SCD risk factors such as syncope and family history of SCD (FH-SCD) as well as family history of HCM (FH-HCM) are documented in electronic health records (EHRs) as clinical narratives. Automated extraction of risk factors from clinical narratives by natural language processing (NLP) may expedite management workflow of HCM patients. The aim of this study was to develop and deploy NLP algorithms for automated extraction of syncope, FH-SCD, and FH-HCM from clinical narratives.
Methods and results: We randomly selected 200 patients from the Mayo HCM registry for development (n = 100) and testing (n = 100) of NLP algorithms for extraction of syncope, FH-SCD as well as FH-HCM from clinical narratives of EHRs. The clinical reference standard was manually abstracted by 2 independent annotators. Performance of NLP algorithms was compared to aggregation and summarization of data entries in the HCM registry for syncope, FH-SCD, and FH-HCM. We also compared the NLP algorithms with billing codes for syncope as well as responses to patient survey questions for FH-SCD and FH-HCM. These analyses demonstrated NLP had superior sensitivity (0.96 vs 0.39, p < 0.001) and comparable specificity (0.90 vs 0.92, p = 0.74) and PPV (0.90 vs 0.83, p = 0.37) compared to billing codes for syncope. For FH-SCD, NLP outperformed survey responses for all parameters (sensitivity: 0.91 vs 0.59, p = 0.002; specificity: 0.98 vs 0.50, p < 0.001; PPV: 0.97 vs 0.38, p < 0.001). NLP also achieved superior sensitivity (0.95 vs 0.24, p < 0.001) with comparable specificity (0.95 vs 1.0, p-value not calculable) and positive predictive value (PPV) (0.92 vs 1.0, p = 0.09) compared to survey responses for FH-HCM.
Conclusions: Automated extraction of syncope, FH-SCD and FH-HCM using NLP is feasible and has promise to increase efficiency of workflow for providers managing HCM patients.
Keywords: Electronic health records; Hypertrophic cardiomyopathy; Natural language processing; Sudden cardiac death.
Copyright © 2019 The Authors. Published by Elsevier B.V. All rights reserved.
Figures
Similar articles
-
Sudden cardiac death in patients with hypertrophic cardiomyopathy: from bench to bedside with an emphasis on genetic markers.Clin Cardiol. 1995 Apr;18(4):189-98. doi: 10.1002/clc.4960180403. Clin Cardiol. 1995. PMID: 7788945 Review.
-
A validation study of the 2003 American College of Cardiology/European Society of Cardiology and 2011 American College of Cardiology Foundation/American Heart Association risk stratification and treatment algorithms for sudden cardiac death in patients with hypertrophic cardiomyopathy.Heart. 2013 Apr;99(8):534-41. doi: 10.1136/heartjnl-2012-303271. Epub 2013 Jan 22. Heart. 2013. PMID: 23339826
-
A novel clinical risk prediction model for sudden cardiac death in hypertrophic cardiomyopathy (HCM risk-SCD).Eur Heart J. 2014 Aug 7;35(30):2010-20. doi: 10.1093/eurheartj/eht439. Epub 2013 Oct 14. Eur Heart J. 2014. PMID: 24126876
-
Assessment of the relationship between the ambulatory electrocardiography-based micro T-wave alternans and the predicted risk score of sudden cardiac death at 5 years in patients with hypertrophic cardiomyopathy.Anatol J Cardiol. 2018 Sep;20(3):165-173. doi: 10.14744/AnatolJCardiol.2018.15945. Anatol J Cardiol. 2018. PMID: 30152798 Free PMC article.
-
Risk factors of sudden cardiac death in hypertrophic cardiomyopathy.Curr Opin Cardiol. 2022 Jan 1;37(1):15-21. doi: 10.1097/HCO.0000000000000939. Curr Opin Cardiol. 2022. PMID: 34636345 Free PMC article. Review.
Cited by
-
Large Language Models for Efficient Medical Information Extraction.AMIA Jt Summits Transl Sci Proc. 2024 May 31;2024:509-514. eCollection 2024. AMIA Jt Summits Transl Sci Proc. 2024. PMID: 38827084 Free PMC article.
-
Conversion of left atrial volume to diameter for automated estimation of sudden cardiac death risk in hypertrophic cardiomyopathy.Echocardiography. 2021 Feb;38(2):183-188. doi: 10.1111/echo.14943. Epub 2020 Dec 16. Echocardiography. 2021. PMID: 33325582 Free PMC article.
-
The path from big data analytics capabilities to value in hospitals: a scoping review.BMC Health Serv Res. 2022 Jan 31;22(1):134. doi: 10.1186/s12913-021-07332-0. BMC Health Serv Res. 2022. PMID: 35101026 Free PMC article.
-
Automated Identification of Aspirin-Exacerbated Respiratory Disease Using Natural Language Processing and Machine Learning: Algorithm Development and Evaluation Study.JMIR AI. 2023 Jun 12;2:e44191. doi: 10.2196/44191. JMIR AI. 2023. PMID: 39105270 Free PMC article.
-
Artificial Intelligence and Cardiovascular Genetics.Life (Basel). 2022 Feb 14;12(2):279. doi: 10.3390/life12020279. Life (Basel). 2022. PMID: 35207566 Free PMC article. Review.
References
-
- Jensen PB, Jensen LJ and Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012;13:395–405. - PubMed
-
- Maddox TM, Albert NM, Borden WB, Curtis LH, Ferguson TB Jr., Kao DP, Marcus GM, Peterson ED, Redberg R, Rumsfeld JS, Shah ND, Tcheng JE, American Heart Association Council on Quality of C, Outcomes R, Council on Cardiovascular Disease in the Y, Council on Clinical C, Council on Functional G, Translational B and Stroke C. The Learning Healthcare System and Cardiovascular Care: A Scientific Statement From the American Heart Association. Circulation. 2017;135:e826–e857. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

