Growth disrupting mutations in epigenetic regulatory molecules are associated with abnormalities of epigenetic aging
- PMID: 31160375
- PMCID: PMC6633263
- DOI: 10.1101/gr.243584.118
Growth disrupting mutations in epigenetic regulatory molecules are associated with abnormalities of epigenetic aging
Abstract
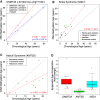
Germline mutations in fundamental epigenetic regulatory molecules including DNA methyltransferase 3 alpha (DNMT3A) are commonly associated with growth disorders, whereas somatic mutations are often associated with malignancy. We profiled genome-wide DNA methylation patterns in DNMT3A c.2312G > A; p.(Arg771Gln) carriers in a large Amish sibship with Tatton-Brown-Rahman syndrome (TBRS), their mosaic father, and 15 TBRS patients with distinct pathogenic de novo DNMT3A variants. This defined widespread DNA hypomethylation at specific genomic sites enriched at locations annotated as genes involved in morphogenesis, development, differentiation, and malignancy predisposition pathways. TBRS patients also displayed highly accelerated DNA methylation aging. These findings were most marked in a carrier of the AML-associated driver mutation p.Arg882Cys. Our studies additionally defined phenotype-related accelerated and decelerated epigenetic aging in two histone methyltransferase disorders: NSD1 Sotos syndrome overgrowth disorder and KMT2D Kabuki syndrome growth impairment. Together, our findings provide fundamental new insights into aberrant epigenetic mechanisms, the role of epigenetic machinery maintenance, and determinants of biological aging in these growth disorders.
© 2019 Jeffries et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57: 289–300. 10.2307/2346101 - DOI
-
- Butcher DT, Cytrynbaum C, Turinsky AL, Siu MT, Inbar-Feigenberg M, Mendoza-Londono R, Chitayat D, Walker S, Machado J, Caluseriu O, et al. 2017. CHARGE and Kabuki syndromes: Gene-specific DNA methylation signatures identify epigenetic mechanisms linking these clinically overlapping conditions. Am J Hum Genet 100: 773–788. 10.1016/j.ajhg.2017.04.004 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
