Tfap2a is a novel gatekeeper of nephron differentiation during kidney development
- PMID: 31160420
- PMCID: PMC6633607
- DOI: 10.1242/dev.172387
Tfap2a is a novel gatekeeper of nephron differentiation during kidney development
Abstract
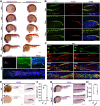
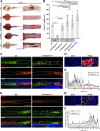
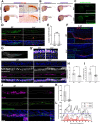
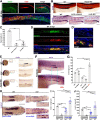
Renal functional units known as nephrons undergo patterning events during development that create a segmental array of cellular compartments with discrete physiological identities. Here, from a forward genetic screen using zebrafish, we report the discovery that transcription factor AP-2 alpha (tfap2a) coordinates a gene regulatory network that activates the terminal differentiation program of distal segments in the pronephros. We found that tfap2a acts downstream of Iroquois homeobox 3b (irx3b), a distal lineage transcription factor, to operate a circuit consisting of tfap2b, irx1a and genes encoding solute transporters that dictate the specialized metabolic functions of distal nephron segments. Interestingly, this regulatory node is distinct from other checkpoints of differentiation, such as polarity establishment and ciliogenesis. Thus, our studies reveal insights into the genetic control of differentiation, where tfap2a is essential for regulating a suite of segment transporter traits at the final tier of zebrafish pronephros ontogeny. These findings have relevance for understanding renal birth defects, as well as efforts to recapitulate nephrogenesis in vivo to facilitate drug discovery and regenerative therapies.
Keywords: Differentiation; Kidney; Nephron; Segmentation; Zebrafish; irx1a; irx3b; tfap2a; tfap2b.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

