A comprehensive single cell transcriptional landscape of human hematopoietic progenitors
- PMID: 31160568
- PMCID: PMC6546699
- DOI: 10.1038/s41467-019-10291-0
A comprehensive single cell transcriptional landscape of human hematopoietic progenitors
Abstract
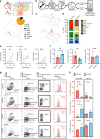
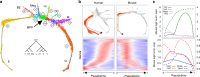
Hematopoietic Stem/Progenitor cells (HSPCs) are endowed with the role of maintaining a diverse pool of blood cells throughout the human life. Despite recent efforts, the nature of the early cell fate decisions remains contentious. Using single-cell RNA-Seq, we show that existing approaches to stratify bone marrow CD34+ cells reveal a hierarchically-structured transcriptional landscape of hematopoietic differentiation. Still, this landscape misses important early fate decisions. We here provide a broader transcriptional profiling of bone marrow lineage negative hematopoietic progenitors that recovers a key missing branchpoint into basophils and expands our understanding of the underlying structure of early adult human haematopoiesis. We also show that this map has strong similarities in topology and gene expression to that found in mouse. Finally, we identify the sialomucin CD164, as a reliable marker for the earliest branches of HSPCs specification and we showed how its use can foster the design of alternative transplantation cell products.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

