Complex N-glycan breakdown by gut Bacteroides involves an extensive enzymatic apparatus encoded by multiple co-regulated genetic loci
- PMID: 31160824
- PMCID: PMC7617214
- DOI: 10.1038/s41564-019-0466-x
Complex N-glycan breakdown by gut Bacteroides involves an extensive enzymatic apparatus encoded by multiple co-regulated genetic loci
Abstract
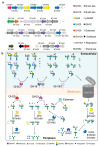
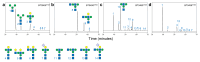
Glycans are the major carbon sources available to the human colonic microbiota. Numerous N-glycosylated proteins are found in the human gut, from both dietary and host sources, including immunoglobulins such as IgA that are secreted into the intestine at high levels. Here, we show that many mutualistic gut Bacteroides spp. have the capacity to utilize complex N-glycans (CNGs) as nutrients, including those from immunoglobulins. Detailed mechanistic studies using transcriptomic, biochemical, structural and genetic techniques reveal the pathway employed by Bacteroides thetaiotaomicron (Bt) for CNG degradation. The breakdown process involves an extensive enzymatic apparatus encoded by multiple non-adjacent loci and comprises 19 different carbohydrate-active enzymes from different families, including a CNG-specific endo-glycosidase activity. Furthermore, CNG degradation involves the activity of carbohydrate-active enzymes that have previously been implicated in the degradation of other classes of glycan. This complex and diverse apparatus provides Bt with the capacity to access the myriad different structural variants of CNGs likely to be found in the intestinal niche.
Conflict of interest statement
The authors declare no competing interests
Figures




References
-
- McNeil NI. The contribution of the large intestine to energy supplies in man. The American journal of clinical nutrition. 1984;39:338–342. - PubMed
-
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. - PubMed
-
- Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. Journal of clinical gastroenterology. 2010;44:354–360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

