Genetic population structure and demography of an apex predator, the tiger shark Galeocerdo cuvier
- PMID: 31160982
- PMCID: PMC6540675
- DOI: 10.1002/ece3.5111
Genetic population structure and demography of an apex predator, the tiger shark Galeocerdo cuvier
Abstract
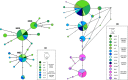
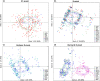
Population genetics has been increasingly applied to study large sharks over the last decade. Whilst large shark species are often difficult to study with direct methods, improved knowledge is needed for both population management and conservation, especially for species vulnerable to anthropogenic and climatic impacts. The tiger shark, Galeocerdo cuvier, is an apex predator known to play important direct and indirect roles in tropical and subtropical marine ecosystems. While the global and Indo-West Pacific population genetic structure of this species has recently been investigated, questions remain over population structure and demographic history within the western Indian (WIO) and within the western Pacific Oceans (WPO). To address the knowledge gap in tiger shark regional population structures, the genetic diversity of 286 individuals sampled in seven localities was investigated using 27 microsatellite loci and three mitochondrial genes (CR,COI, and cytb). A weak genetic differentiation was observed between the WIO and the WPO, suggesting high genetic connectivity. This result agrees with previous studies and highlights the importance of the pelagic behavior of this species to ensure gene flow. Using approximate Bayesian computation to couple information from both nuclear and mitochondrial markers, evidence of a recent bottleneck in the Holocene (2,000-3,000 years ago) was found, which is the most probable cause for the low genetic diversity observed. A contemporary effective population size as low as 111 [43,369] was estimated during the bottleneck. Together, these results indicate low genetic diversity that may reflect a vulnerable population sensitive to regional pressures. Conservation measures are thus needed to protect a species that is classified as Near Threatened.
Keywords: approximate Bayesian computation; bottleneck; effective population size; microsatellite DNA; mitochondrial DNA; tiger shark.
Conflict of interest statement
None declared.
Figures

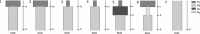

References
-
- Afonso, A. S. , & Hazin, F. H. V. (2014). Post‐release survival and behavior and exposure to fisheries in juvenile tiger sharks, Galeocerdo cuvier, from the South Atlantic. Journal of Experimental Marine Biology and Ecology, 454, 55–62. 10.1016/j.jembe.2014.02.008 - DOI
-
- Aronson, R. B. , & Precht, W. F. (2001). White‐band disease and the changing face of Caribbean coral reefs In Porter J. W. (Ed.), The ecology and etiology of newly emerging marine diseases (pp. 25–38). Dordrecht, the Netherlands: Springer Netherlands; 10.1007/978-94-017-3284-0_2 - DOI
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

