Temporal Stability and Prognostic Biomarker Potential of the Prostate Cancer Urine miRNA Transcriptome
- PMID: 31161221
- PMCID: PMC7073919
- DOI: 10.1093/jnci/djz112
Temporal Stability and Prognostic Biomarker Potential of the Prostate Cancer Urine miRNA Transcriptome
Abstract
Background: The development of noninvasive tests for the early detection of aggressive prostate tumors is a major unmet clinical need. miRNAs are promising noninvasive biomarkers: they play essential roles in tumorigenesis, are stable under diverse analytical conditions, and can be detected in body fluids.
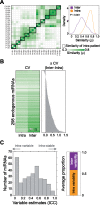
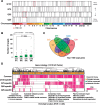
Methods: We measured the longitudinal stability of 673 miRNAs by collecting serial urine samples from 10 patients with localized prostate cancer. We then measured temporally stable miRNAs in an independent training cohort (n = 99) and created a biomarker predictive of Gleason grade using machine-learning techniques. Finally, we validated this biomarker in an independent validation cohort (n = 40).
Results: We found that each individual has a specific urine miRNA fingerprint. These fingerprints are temporally stable and associated with specific biological functions. We identified seven miRNAs that were stable over time within individual patients and integrated them with machine-learning techniques to create a novel biomarker for prostate cancer that overcomes interindividual variability. Our urine biomarker robustly identified high-risk patients and achieved similar accuracy as tissue-based prognostic markers (area under the receiver operating characteristic = 0.72, 95% confidence interval = 0.69 to 0.76 in the training cohort, and area under the receiver operating characteristic curve = 0.74, 95% confidence interval = 0.55 to 0.92 in the validation cohort).
Conclusions: These data highlight the importance of quantifying intra- and intertumoral heterogeneity in biomarker development. This noninvasive biomarker may usefully supplement invasive or expensive radiologic- and tissue-based assays.
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures


Comment in
-
Holding a MIRror up to the robustness of the prostate cancer urinary transcriptome.Transl Androl Urol. 2019 Dec;8(Suppl 5):S488-S490. doi: 10.21037/tau.2019.09.25. Transl Androl Urol. 2019. PMID: 32042627 Free PMC article. No abstract available.
References
-
- Siegel R, Naishadham D, Jemal A.. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. - PubMed
-
- Musunuru HB, Yamamoto T, Klotz L, et al. Active surveillance for intermediate risk prostate cancer: survival outcomes in the Sunnybrook experience. J Urol. 2016;196(6):1651–1658. - PubMed
-
- Loeb S, Vellekoop A, Ahmed HU, et al. Systematic review of complications of prostate biopsy. Eur Urol. 2013;64(6):876–892. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

