BAL Cell Gene Expression in Severe Asthma Reveals Mechanisms of Severe Disease and Influences of Medications
- PMID: 31161938
- PMCID: PMC6812436
- DOI: 10.1164/rccm.201811-2221OC
BAL Cell Gene Expression in Severe Asthma Reveals Mechanisms of Severe Disease and Influences of Medications
Abstract
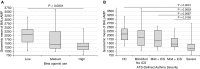
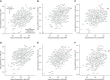
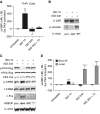
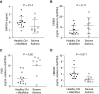
Rationale: Gene expression of BAL cells, which samples the cellular milieu within the lower respiratory tract, has not been well studied in severe asthma.Objectives: To identify new biomolecular mechanisms underlying severe asthma by an unbiased, detailed interrogation of global gene expression.Methods: BAL cell expression was profiled in 154 asthma and control subjects. Of these participants, 100 had accompanying airway epithelial cell gene expression. BAL cell expression profiles were related to participant (age, sex, race, and medication) and sample traits (cell proportions), and then severity-related gene expression determined by correlating transcripts and coexpression networks to lung function, emergency department visits or hospitalizations in the last year, medication use, and quality-of-life scores.Measurements and Main Results: Age, sex, race, cell proportions, and medications strongly influenced BAL cell gene expression, but leading severity-related genes could be determined by carefully identifying and accounting for these influences. A BAL cell expression network enriched for cAMP signaling components most differentiated subjects with severe asthma from other subjects. Subsequently, an in vitro cellular model showed this phenomenon was likely caused by a robust upregulation in cAMP-related expression in nonsevere and β-agonist-naive subjects given a β-agonist before cell collection. Interestingly, ELISAs performed on BAL lysates showed protein levels may partly disagree with expression changes.Conclusions: Gene expression in BAL cells is influenced by factors seldomly considered. Notably, β-agonist exposure likely had a strong and immediate impact on cellular gene expression, which may not translate to important disease mechanisms or necessarily match protein levels. Leading severity-related genes were discovered in an unbiased, system-wide analysis, revealing new targets that map to asthma susceptibility loci.
Keywords: asthma; bronchoalveolar lavage; gene expression; genetics; β-agonist.
Figures






Comment in
-
Advancing Understanding of Mechanisms of Severe Asthma and Drug Effects Using Transcriptomics.Am J Respir Crit Care Med. 2019 Oct 1;200(7):795-796. doi: 10.1164/rccm.201905-0953ED. Am J Respir Crit Care Med. 2019. PMID: 31166693 Free PMC article. No abstract available.
References
-
- Sullivan SD, Rasouliyan L, Russo PA, Kamath T, Chipps BE TENOR Study Group. Extent, patterns, and burden of uncontrolled disease in severe or difficult-to-treat asthma. Allergy. 2007;62:126–133. - PubMed
-
- Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18:716–725. - PubMed
-
- Cookson W. The alliance of genes and environment in asthma and allergy. Nature. 1999;402(Suppl):B5–B11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K08 HL126135/HL/NHLBI NIH HHS/United States
- R01 HL069174/HL/NHLBI NIH HHS/United States
- R01 HL069116/HL/NHLBI NIH HHS/United States
- K23 HL144418/HL/NHLBI NIH HHS/United States
- R03 HL144888/HL/NHLBI NIH HHS/United States
- P01 HL103453/HL/NHLBI NIH HHS/United States
- R01 HL069167/HL/NHLBI NIH HHS/United States
- UL1 RR024153/RR/NCRR NIH HHS/United States
- RC2 HL101487/HL/NHLBI NIH HHS/United States
- U10 HL109250/HL/NHLBI NIH HHS/United States
- UL1 RR025011/RR/NCRR NIH HHS/United States
- P01 AI106684/AI/NIAID NIH HHS/United States
- UL1 TR002373/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

