Discovering sequence and structure landscapes in RNA interaction motifs
- PMID: 31162604
- PMCID: PMC6547422
- DOI: 10.1093/nar/gkz250
Discovering sequence and structure landscapes in RNA interaction motifs
Abstract
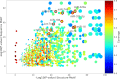
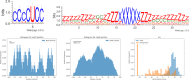
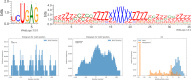
RNA molecules are able to bind proteins, DNA and other small or long RNAs using information at primary, secondary or tertiary structure level. Recent techniques that use cross-linking and immunoprecipitation of RNAs can detect these interactions and, if followed by high-throughput sequencing, molecules can be analysed to find recurrent elements shared by interactors, such as sequence and/or structure motifs. Many tools are able to find sequence motifs from lists of target RNAs, while others focus on structure using different approaches to find specific interaction elements. In this work, we make a systematic analysis of RBP-RNA and RNA-RNA datasets to better characterize the interaction landscape with information about multi-motifs on the same RNAs. To achieve this goal, we updated our BEAM algorithm to combine both sequence and structure information to create pairs of patterns that model motifs of interaction. This algorithm was applied to several RNA binding proteins and ncRNAs interactors, confirming already known motifs and discovering new ones. This landscape analysis on interaction variability reflects the diversity of target recognition and underlines that often both primary and secondary structure are involved in molecular recognition.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
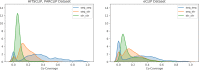
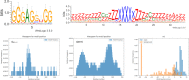
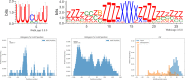
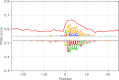
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

