Delivery of self-amplifying RNA vaccines in in vitro reconstituted virus-like particles
- PMID: 31163034
- PMCID: PMC6548422
- DOI: 10.1371/journal.pone.0215031
Delivery of self-amplifying RNA vaccines in in vitro reconstituted virus-like particles
Abstract
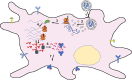
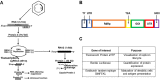
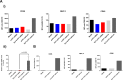
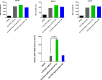
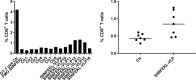
Many mRNA-based vaccines have been investigated for their specific potential to activate dendritic cells (DCs), the highly-specialized antigen-presenting cells of the immune system that play a key role in inducing effective CD4+ and CD8+ T-cell responses. In this paper we report a new vaccine/gene delivery platform that demonstrates the benefits of using a self-amplifying ("replicon") mRNA that is protected in a viral-protein capsid. Purified capsid protein from the plant virus Cowpea Chlorotic Mottle Virus (CCMV) is used to in vitro assemble monodisperse virus-like particles (VLPs) containing reporter proteins (e.g., Luciferase or eYFP) or the tandem-repeat model antigen SIINFEKL in RNA gene form, coupled to the RNA-dependent RNA polymerase from the Nodamura insect virus. Incubation of immature DCs with these VLPs results in increased activation of maturation markers - CD80, CD86 and MHC-II - and enhanced RNA replication levels, relative to incubation with unpackaged replicon mRNA. Higher RNA uptake/replication and enhanced DC activation were detected in a dose-dependent manner when the CCMV-VLPs were pre-incubated with anti-CCMV antibodies. In all experiments the expression of maturation markers correlates with the RNA levels of the DCs. Overall, these studies demonstrate that: VLP protection enhances mRNA uptake by DCs; coupling replicons to the gene of interest increases RNA and protein levels in the cell; and the presence of anti-VLP antibodies enhances mRNA levels and activation of DCs in vitro. Finally, preliminary in vivo experiments involving mouse vaccinations with SIINFEKL-replicon VLPs indicate a small but significant increase in antigen-specific T cells that are doubly positive for IFN and TFN induction.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Chahal JS, Khan OF, Cooper L, McPartlan JS, Tsossie JK, Tilley LD, et al. Dendrimer-RNA nanoparticles generate protective immunity against lethal ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc. Nat. Acad. Sci. USA 2016; 113: E4133–42. 10.1073/pnas.1600299113 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

