Expression Pattern of FT/TFL1 and miR156-Targeted SPL Genes Associated with Developmental Stages in Dendrobium catenatum
- PMID: 31163611
- PMCID: PMC6600168
- DOI: 10.3390/ijms20112725
Expression Pattern of FT/TFL1 and miR156-Targeted SPL Genes Associated with Developmental Stages in Dendrobium catenatum
Abstract
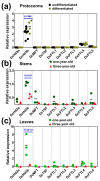
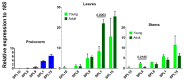
Time to flower, a process either referring to juvenile-adult phase change or vegetative-reproductive transition, is strictly controlled by an intricate regulatory network involving at least both FT/TFL1 and the micro RNA (miR)156-regulated SPL family members. Despite substantial progresses recently achieved in Arabidopsis and other plant species, information regarding the involvement of these genes during orchid development and flowering competence is still limited. Dendrobium catenatum, a popular orchid species, exhibits a juvenile phase of at least three years. Here, through whole-genome mining and whole-family expression profiling, we analyzed the homologous genes of FT/TFL1, miR156, and SPL with special reference to the developmental stages. The FT/TFL1 family contains nine members; among them, DcHd3b transcribes abundantly in young and juvenile tissues but not in adult, contrasting with the low levels of others. We also found that mature miR156, encoded by a single locus, accumulated in large quantity in protocorms and declined by seedling development, coincident with an increase in transcripts of three of its targeted SPL members, namely DcSPL14, DcSPL7, and DcSPL18. Moreover, among the seven predicted miR156-targeted SPLs, only DcSPL3 was significantly expressed in adult plants and was associated with plant maturation. Our results might suggest that the juvenile phase change or maturation in this orchid plant likely involves both the repressive action of a TFL1-like pathway and the promotive effect from an SPL3-mediated mechanism.
Keywords: Dendrobium catenatum; TFL1-like; flowering; juvenile; miR156; phosphatidylethanolamine-binding protein (PEBP) family; squamosa promoter binding protein-like (SPL) transcription factor.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Developmental Functions of miR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Genes in Arabidopsis thaliana.PLoS Genet. 2016 Aug 19;12(8):e1006263. doi: 10.1371/journal.pgen.1006263. eCollection 2016 Aug. PLoS Genet. 2016. PMID: 27541584 Free PMC article.
-
Revisiting the phosphatidylethanolamine-binding protein (PEBP) gene family reveals cryptic FLOWERING LOCUS T gene homologs in gymnosperms and sheds new light on functional evolution.New Phytol. 2016 Nov;212(3):730-744. doi: 10.1111/nph.14066. Epub 2016 Jul 4. New Phytol. 2016. PMID: 27375201
-
The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis.Plant Physiol. 2012 May;159(1):461-78. doi: 10.1104/pp.111.192369. Epub 2012 Mar 16. Plant Physiol. 2012. PMID: 22427344 Free PMC article.
-
A Regulatory Network for miR156-SPL Module in Arabidopsis thaliana.Int J Mol Sci. 2019 Dec 6;20(24):6166. doi: 10.3390/ijms20246166. Int J Mol Sci. 2019. PMID: 31817723 Free PMC article. Review.
-
The miR156/SPL Module, a Regulatory Hub and Versatile Toolbox, Gears up Crops for Enhanced Agronomic Traits.Mol Plant. 2015 May;8(5):677-88. doi: 10.1016/j.molp.2015.01.008. Epub 2015 Jan 21. Mol Plant. 2015. PMID: 25617719 Review.
Cited by
-
Sugar Signaling and Post-transcriptional Regulation in Plants: An Overlooked or an Emerging Topic?Front Plant Sci. 2020 Nov 5;11:578096. doi: 10.3389/fpls.2020.578096. eCollection 2020. Front Plant Sci. 2020. PMID: 33224165 Free PMC article. Review.
-
Alterations in Growth Habit to Channel End-of-Season Perennial Reserves towards Increased Yield and Reduced Regrowth after Defoliation in Upland Cotton (Gossypium hirsutum L.).Int J Mol Sci. 2023 Sep 16;24(18):14174. doi: 10.3390/ijms241814174. Int J Mol Sci. 2023. PMID: 37762483 Free PMC article.
-
Characterizing the Role of the miR156-SPL Network in Plant Development and Stress Response.Plants (Basel). 2020 Sep 15;9(9):1206. doi: 10.3390/plants9091206. Plants (Basel). 2020. PMID: 32942558 Free PMC article. Review.
-
Regulatory frameworks involved in the floral induction, formation and developmental programming of woody horticultural plants: a case study on blueberries.Front Plant Sci. 2024 Feb 12;15:1336892. doi: 10.3389/fpls.2024.1336892. eCollection 2024. Front Plant Sci. 2024. PMID: 38410737 Free PMC article. Review.
-
Unveiling the phenology and associated floral regulatory pathways of Humulus lupulus L. in subtropical conditions.Planta. 2024 May 10;259(6):150. doi: 10.1007/s00425-024-04428-9. Planta. 2024. PMID: 38727772
References
-
- Hackett W.P. Juvenility, maturation and rejuvenation in woody plants. Hortic. Rev. 1985;7:109–155.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

