Development and Optimization of a Miniaturized Western Blot-Based Screening Platform to Identify Regulators of Post-Translational Modifications
- PMID: 31163614
- PMCID: PMC6631403
- DOI: 10.3390/ht8020015
Development and Optimization of a Miniaturized Western Blot-Based Screening Platform to Identify Regulators of Post-Translational Modifications
Abstract
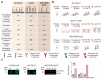
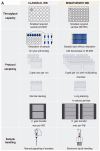
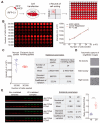
Post-translational modifications (PTMs) are fundamental traits of protein functionality and their study has been addressed using several approaches over the past years. However, screening methods developed to detect regulators of PTMs imply many challenges and are usually based on expensive techniques. Herein, we described the development and optimization of a western blot-based platform for identification of regulators of a specific PTM-mono-ubiquitylation of proliferating cell nuclear antigen (PCNA). This cell-based method does not require specific equipment, apart from the basic western blot (WB) devices and minor accessories, which are accessible for most research labs. The modifications introduced to the classical WB protocol allow the performance of PTM analysis from a single well of a 96-well plate with minimal sample manipulation and low intra- and inter-plate variability, making this method ideal to screen arrayed compound libraries in a 96-well format. As such, our experimental pipeline provides the proof of concept to design small screenings of PTM regulators by improving the quantitative accuracy and throughput capacity of classical western blots.
Keywords: post-translational modification; screening; western blot.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
Similar articles
-
Sensitive Western-Blot Analysis of Azide-Tagged Protein Post Translational Modifications Using Thermoresponsive Polymer Self-Assembly.Anal Chem. 2018 Feb 6;90(3):2186-2192. doi: 10.1021/acs.analchem.7b04531. Epub 2018 Jan 16. Anal Chem. 2018. PMID: 29283250
-
Functional analysis tools for post-translational modification: a post-translational modification database for analysis of proteins and metabolic pathways.Plant J. 2019 Sep;99(5):1003-1013. doi: 10.1111/tpj.14372. Epub 2019 May 31. Plant J. 2019. PMID: 31034103
-
A homology-based pipeline for global prediction of post-translational modification sites.Sci Rep. 2016 May 13;6:25801. doi: 10.1038/srep25801. Sci Rep. 2016. PMID: 27174170 Free PMC article.
-
Enrichment and separation techniques for large-scale proteomics analysis of the protein post-translational modifications.J Chromatogr A. 2014 Dec 12;1372C:1-17. doi: 10.1016/j.chroma.2014.10.107. Epub 2014 Nov 6. J Chromatogr A. 2014. PMID: 25465002 Review.
-
Fishing the PTM proteome with chemical approaches using functional solid phases.Chem Soc Rev. 2015 Nov 21;44(22):8260-87. doi: 10.1039/c4cs00529e. Epub 2015 Aug 10. Chem Soc Rev. 2015. PMID: 26258179 Review.
Cited by
-
Recent Advancements in Electrochemical Biosensors for Alzheimer's Disease Biomarkers Detection.Curr Med Chem. 2021;28(20):4049-4073. doi: 10.2174/0929867327666201111141341. Curr Med Chem. 2021. PMID: 33176635 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

