HHIPL1, a Gene at the 14q32 Coronary Artery Disease Locus, Positively Regulates Hedgehog Signaling and Promotes Atherosclerosis
- PMID: 31163988
- PMCID: PMC6686954
- DOI: 10.1161/CIRCULATIONAHA.119.041059
HHIPL1, a Gene at the 14q32 Coronary Artery Disease Locus, Positively Regulates Hedgehog Signaling and Promotes Atherosclerosis
Abstract
Background: Genome-wide association studies have identified chromosome 14q32 as a locus for coronary artery disease. The disease-associated variants fall in a hitherto uncharacterized gene called HHIPL1 (hedgehog interacting protein-like 1), which encodes a sequence homolog of an antagonist of hedgehog signaling. The function of HHIPL1 and its role in atherosclerosis are unknown.
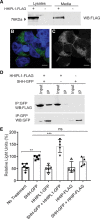
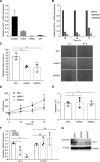
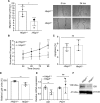
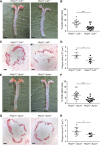
Methods: HHIPL1 cellular localization, interaction with sonic hedgehog (SHH), and influence on hedgehog signaling were tested. HHIPL1 expression was measured in coronary artery disease-relevant human cells, and protein localization was assessed in wild-type and Apoe-/- (apolipoprotein E deficient) mice. Human aortic smooth muscle cell phenotypes and hedgehog signaling were investigated after gene knockdown. Hhipl1-/- mice were generated and aortic smooth muscle cells collected for phenotypic analysis and assessment of hedgehog signaling activity. Hhipl1-/- mice were bred onto both the Apoe-/- and Ldlr-/- (low-density lipoprotein receptor deficient) knockout strains, and the extent of atherosclerosis was quantified after 12 weeks of high-fat diet. Cellular composition and collagen content of aortic plaques were assessed by immunohistochemistry.
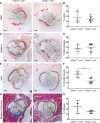
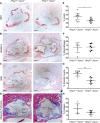
Results: In vitro analyses revealed that HHIPL1 is a secreted protein that interacts with SHH and increases hedgehog signaling activity. HHIPL1 expression was detected in human smooth muscle cells and in smooth muscle within atherosclerotic plaques of Apoe-/- mice. The expression of Hhipl1 increased with disease progression in aortic roots of Apoe-/- mice. Proliferation and migration were reduced in Hhipl1 knockout mouse and HHIPL1 knockdown aortic smooth muscle cells, and hedgehog signaling was decreased in HHIPL1-deficient cells. Hhipl1 knockout caused a reduction of >50% in atherosclerosis burden on both Apoe-/- and Ldlr-/- knockout backgrounds, and lesions were characterized by reduced smooth muscle cell content.
Conclusions: HHIPL1 is a secreted proatherogenic protein that enhances hedgehog signaling and regulates smooth muscle cell proliferation and migration. Inhibition of HHIPL1 protein function might offer a novel therapeutic strategy for coronary artery disease.
Keywords: atherosclerosis; coronary artery disease; genome-wide association study; hedgehogs; signaling.
Figures

References
-
- Schunkert H, König IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, et al. Cardiogenics; CARDIoGRAM Consortium. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. - PMC - PubMed
-
- Klarin D, Zhu QM, Emdin CA, Chaffin M, Horner S, McMillan BJ, Leed A, Weale ME, Spencer CCA, Aguet F, et al. CARDIoGRAMplusC4D Consortium. Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat Genet. 2017;49:1392–1397. doi: 10.1038/ng.3914. - PMC - PubMed
-
- Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, et al. EPIC-CVD Consortium; CARDIoGRAMplusC4D; UK Biobank CardioMetabolic Consortium CHD working group. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913. - PubMed
-
- Katoh Y, Katoh M. Comparative genomics on HHIP family orthologs. Int J Mol Med. 2006;17:391–395. doi: 10.3892/ijmm.17.2.391. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

