Regulation of gene expression in trypanosomatids: living with polycistronic transcription
- PMID: 31164043
- PMCID: PMC6597758
- DOI: 10.1098/rsob.190072
Regulation of gene expression in trypanosomatids: living with polycistronic transcription
Abstract
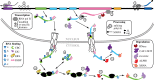
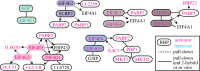
In trypanosomes, RNA polymerase II transcription is polycistronic and individual mRNAs are excised by trans-splicing and polyadenylation. The lack of individual gene transcription control is compensated by control of mRNA processing, translation and degradation. Although the basic mechanisms of mRNA decay and translation are evolutionarily conserved, there are also unique aspects, such as the existence of six cap-binding translation initiation factor homologues, a novel decapping enzyme and an mRNA stabilizing complex that is recruited by RNA-binding proteins. High-throughput analyses have identified nearly a hundred regulatory mRNA-binding proteins, making trypanosomes valuable as a model system to investigate post-transcriptional regulation.
Keywords: Leishmania; Trypanosoma; mRNA decay; mRNA processing; mRNA translation; transcription.
Conflict of interest statement
I declare I have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
