Genomic Evolutionary Patterns of Leiomyosarcoma and Liposarcoma
- PMID: 31164371
- PMCID: PMC7341441
- DOI: 10.1158/1078-0432.CCR-19-0271
Genomic Evolutionary Patterns of Leiomyosarcoma and Liposarcoma
Abstract
Purpose: Leiomyosarcoma and liposarcoma are common subtypes of soft tissue sarcoma (STS). Patients with metastatic leiomyosarcoma or dedifferentiated liposarcoma (DDLPS) typically have worse outcomes compared with localized leiomyosarcoma or well-differentiated liposarcoma (WDLPS). A better understanding of genetic changes between primary/metastatic leiomyosarcoma and between WDLPS/DDLPS may provide insight into their genetic evolution.
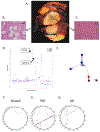
Experimental design: We interrogated whole-exome sequencing (WES) from "trios" of normal tissue, primary tumor, and metastatic tumor from individual patients with leiomyosarcoma (n = 9), and trios of normal tissue, well-differentiated tumor, and dedifferentiated tumor from individual patients with liposarcoma (n = 19). Specifically, we performed mutational, copy number, and tumor evolution analyses on these cohorts and compared patterns among leiomyosarcoma and liposarcoma trios.
Results: Leiomyosarcoma cases harbored shared drivers through a typical parent/child relationship where the metastatic tumor was derived from the primary tumor. In contrast, while all liposarcoma cases shared the characteristic focal chromosome 12 amplicon, most paired liposarcoma cases did not share additional mutations, suggesting a divergent evolutionary pattern from a common precursor. No highly recurrent genomic alterations from WES were identified that could be implicated as driving the progression of disease in either sarcoma subtype.
Conclusions: From a genomic perspective, leiomyosarcoma metastases contain genetic alterations that are also found in primary tumors. WDLPS and DDLPS, however, appear to divergently evolve from a common precursor harboring 12q amplification, rather than as a transformation to a higher-grade tumor. Further efforts to identify specific drivers of these distinct evolutionary patterns may inform future translational and clinical research in STS.
©2019 American Association for Cancer Research.
Figures


References
-
- Serrano C, George S Leiomyosarcoma In: Wagner AJ, editor. Hematology/Oncology Clinics of North America: Sarcoma. Philadelphia: Elsevier, Inc; 2013. p. 957–974. - PubMed
-
- Henze J, Bauer S, Liposarcomas In: Wagner AJ, editor. Hematology/Oncology Clinics of North America: Sarcoma. Philadelphia: Elsevier, Inc; 2013. p. 939–955. - PubMed
-
- Chibon F, Mariani O, Derré J, Mairal A, Coindre JM, Guillou L et al. ASK1 (MAP3K5) as a potential therapeutic target in malignant fibrous histiocytomas with 12q14-q15 and 6q23 amplifications. Genes, Chromosomes & Cancer 40, 32–37 (2004). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

