Molecular Analysis of PKU-Associated PAH Mutations: A Fast and Simple Genotyping Test
- PMID: 31164572
- PMCID: PMC6481045
- DOI: 10.3390/mps1030030
Molecular Analysis of PKU-Associated PAH Mutations: A Fast and Simple Genotyping Test
Abstract
Neonatal screening for phenylketonuria (PKU, OMIM: 261600) was introduced at the end of the 1960s. We developed a rapid and simple molecular test for the most frequent phenylalanine hydroxylase (PAH, Gene ID: 5053) mutations. Using this method to detect the 18 most frequent mutations, it is possible to achieve a 75% detection rate in Italian population. The variants selected also reach a high detection rate in other populations, for example, 70% in southern Germany, 68% in western Germany, 76% in Denmark, 68% in Sweden, 63% in Poland, and 60% in Bulgaria. We successfully applied this confirmation test in neonatal screening for hyperphenylalaninemias using dried blood spots and obtained the genotype in approximately 48 h. The method was found to be suitable as second tier test in neonatal screening for hyperphenylalaninemias in neonates with a positive screening test. This test can also be useful for carrier screening because it can bypass the entire coding sequence and intron-exon boundaries sequencing, thereby overcoming the questions that this approach implies, such as new variant interpretations.
Keywords: PKU mutation analysis; PKU screening; phenylalanine hydroxylase deficiency; phenylketonuria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
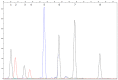

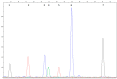
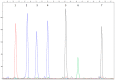

References
-
- Scriver C.R., Kaufman S. The Metabolic and Molecular Bases of Inherited Disease. 8th ed. McGraw-Hill; New York, NY, USA: 2001. The hyperphenylalaninemias; pp. 1667–1724.
-
- Guldberg P., Rey F., Zschocke J., Romano V., François B., Michiels L., Ullrich K., Hoffmann G.F., Burgard P., Schmidt H., et al. A European multicenter study of phenylalanine hydroxylase deficiency: classification of 105 mutations and a general system for genotype-based prediction of metabolic phenotype. Am. J. Hum. Genet. 1998;63:71–79. doi: 10.1086/301920. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

