An analysis of large structural variation in global Plasmodium falciparum isolates identifies a novel duplication of the chloroquine resistance associated gene
- PMID: 31164664
- PMCID: PMC6547842
- DOI: 10.1038/s41598-019-44599-0
An analysis of large structural variation in global Plasmodium falciparum isolates identifies a novel duplication of the chloroquine resistance associated gene
Abstract
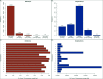
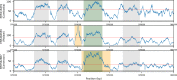
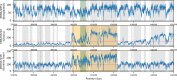
The evolution of genetic mechanisms for host immune evasion and anti-malarial resistance has enabled the Plasmodium falciparum malaria parasite to inflict high morbidity and mortality on human populations. Most studies of P. falciparum genetic diversity have focused on single-nucleotide polymorphisms (SNPs), assisting the identification of drug resistance-associated loci such as the chloroquine related crt and sulfadoxine-pyrimethamine related dhfr. Whilst larger structural variants are known to impact adaptation, for example, mdr1 duplications with anti-malarial resistance, no large-scale, genome-wide study on clinical isolates has been undertaken using whole genome sequencing data. By applying a structural variant detection pipeline across whole genome sequence data from 2,855 clinical isolates in 21 malaria-endemic countries, we identified >70,000 specific deletions and >600 duplications. Most structural variants are rare (48.5% of deletions and 94.7% of duplications are found in single isolates) with 2.4% of deletions and 0.2% of duplications found in >5% of global isolates. A subset of variants was present at high frequency in drug-resistance related genes including mdr1, the gch1 promoter region, and a putative novel duplication of crt. Regional-specific variants were identified, and a companion visualisation tool has been developed to assist web-based investigation of these polymorphisms by the wider scientific community.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- World Health Organization. World Malaria Report 2016. WHO Press (WHO Press, 2016).
Publication types
MeSH terms
Substances
Grants and funding
- MR/K000551/1/MRC_/Medical Research Council/United Kingdom
- MR/M01360X/1/MRC_/Medical Research Council/United Kingdom
- MR/R020973/1/MRC_/Medical Research Council/United Kingdom
- BB/J014567/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/N010469/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Research Materials

