Learning Causal Biological Networks With the Principle of Mendelian Randomization
- PMID: 31164902
- PMCID: PMC6536645
- DOI: 10.3389/fgene.2019.00460
Learning Causal Biological Networks With the Principle of Mendelian Randomization
Abstract
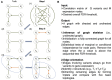
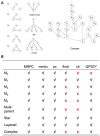
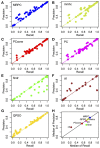
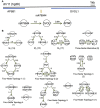
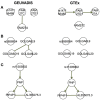
Although large amounts of genomic data are available, it remains a challenge to reliably infer causal (i. e., regulatory) relationships among molecular phenotypes (such as gene expression), especially when multiple phenotypes are involved. We extend the interpretation of the Principle of Mendelian randomization (PMR) and present MRPC, a novel machine learning algorithm that incorporates the PMR in the PC algorithm, a classical algorithm for learning causal graphs in computer science. MRPC learns a causal biological network efficiently and robustly from integrating individual-level genotype and molecular phenotype data, in which directed edges indicate causal directions. We demonstrate through simulation that MRPC outperforms several popular general-purpose network inference methods and PMR-based methods. We apply MRPC to distinguish direct and indirect targets among multiple genes associated with expression quantitative trait loci. Our method is implemented in the R package MRPC, available on CRAN (https://cran.r-project.org/web/packages/MRPC/index.html).
Keywords: Mendelian randomization; bioinformatics; biological networks; causal inference; graphical models.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources

