Protein kinase A inhibits tumor mutator APOBEC3B through phosphorylation
- PMID: 31165764
- PMCID: PMC6549188
- DOI: 10.1038/s41598-019-44407-9
Protein kinase A inhibits tumor mutator APOBEC3B through phosphorylation
Abstract
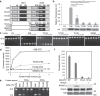
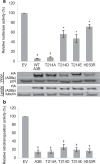
APOBEC3B cytidine deaminase (A3B) catalyzes cytosine into uracil in single-strand DNA and induces C-to-T mutations in genomic DNA of various types of tumors. Accumulation of APOBEC signature mutations is correlated with a worse prognosis for patients with breast cancer or multiple myeloma, suggesting that A3B activity might be a cause of the unfavorable DNA mutations and clonal evolution in these tumors. Phosphorylation of conserved threonine residues of other cytidine deaminases, activation induced deaminase (AID) and APOBEC3G, inhibits their activity. Here we show that protein kinase A (PKA) physically binds to A3B and phosphorylates Thr214. In vitro deaminase assays and foreign DNA editing assays in cells confirm that phosphomimetic A3B mutants, T214D and T214E, completely lose deaminase activity. Molecular dynamics simulation of A3B phosphorylation reveals that Thr214 phosphorylation disrupts binding between the phospho-A3B catalytic core and ssDNA. These mutants still inhibit retroviral infectivity at least partially, and also retain full anti-retrotransposition activity. These results imply that PKA-mediated phosphorylation inhibits A3B mutagenic activity without destructing its innate immune functions. Therefore, PKA activation could reduce further accumulation of mutations in A3B overexpressing tumors.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
The DNA deaminase APOBEC3B interacts with the cell-cycle protein CDK4 and disrupts CDK4-mediated nuclear import of Cyclin D1.J Biol Chem. 2019 Aug 9;294(32):12099-12111. doi: 10.1074/jbc.RA119.008443. Epub 2019 Jun 19. J Biol Chem. 2019. PMID: 31217276 Free PMC article.
-
Mutation Processes in 293-Based Clones Overexpressing the DNA Cytosine Deaminase APOBEC3B.PLoS One. 2016 May 10;11(5):e0155391. doi: 10.1371/journal.pone.0155391. eCollection 2016. PLoS One. 2016. PMID: 27163364 Free PMC article.
-
Endogenous APOBEC3B Overexpression Constitutively Generates DNA Substitutions and Deletions in Myeloma Cells.Sci Rep. 2019 May 9;9(1):7122. doi: 10.1038/s41598-019-43575-y. Sci Rep. 2019. PMID: 31073151 Free PMC article.
-
APOBEC3B: pathological consequences of an innate immune DNA mutator.Biomed J. 2015 Mar-Apr;38(2):102-10. doi: 10.4103/2319-4170.148904. Biomed J. 2015. PMID: 25566802 Review.
-
Unfavorable prognosis and clinical consequences of APOBEC3B expression in breast and other cancers: A systematic review and meta-analysis.Tumour Biol. 2022;44(1):153-169. doi: 10.3233/TUB-211577. Tumour Biol. 2022. PMID: 36093650
Cited by
-
ILF2 enhances the DNA cytosine deaminase activity of tumor mutator APOBEC3B in multiple myeloma cells.Sci Rep. 2022 Feb 10;12(1):2278. doi: 10.1038/s41598-022-06226-3. Sci Rep. 2022. PMID: 35145187 Free PMC article.
-
APOBEC3A is a prominent cytidine deaminase in breast cancer.PLoS Genet. 2019 Dec 16;15(12):e1008545. doi: 10.1371/journal.pgen.1008545. eCollection 2019 Dec. PLoS Genet. 2019. PMID: 31841499 Free PMC article.
-
Molecular origins of APOBEC-associated mutations in cancer.DNA Repair (Amst). 2020 Oct;94:102905. doi: 10.1016/j.dnarep.2020.102905. Epub 2020 Jul 6. DNA Repair (Amst). 2020. PMID: 32818816 Free PMC article. Review.
-
Cancer Evo-Dev: A Theory of Inflammation-Induced Oncogenesis.Front Immunol. 2021 Nov 22;12:768098. doi: 10.3389/fimmu.2021.768098. eCollection 2021. Front Immunol. 2021. PMID: 34880864 Free PMC article.
-
Alternative Splicing, RNA Editing, and the Current Limits of Next Generation Sequencing.Genes (Basel). 2023 Jun 30;14(7):1386. doi: 10.3390/genes14071386. Genes (Basel). 2023. PMID: 37510291 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

