The IL-33/ST2 pathway shapes the regulatory T cell phenotype to promote intestinal cancer
- PMID: 31165767
- PMCID: PMC7746527
- DOI: 10.1038/s41385-019-0176-y
The IL-33/ST2 pathway shapes the regulatory T cell phenotype to promote intestinal cancer
Abstract
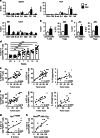
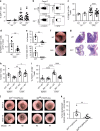
The composition of immune infiltrates strongly affects the prognosis of patients with colorectal cancer (CRC). Interleukin (IL)-33 and regulatory T cells (Tregs) in the tumor microenvironment have been separately implicated in CRC; however their contribution to intestinal carcinogenesis is still controversial. Here, we reveal that IL-33 signaling promotes CRC by changing the phenotype of Tregs. In mice with CRC, tumor-infiltrating Tregs preferentially upregulate IL-33 receptor (ST2), and IL-33/ST2 signaling positively correlates with tumor number and size. Transcriptomic and flow cytometry analyses demonstrate that ST2 expression induces a more activated and migratory phenotype in FOXP3+ Tregs, which favors their accumulation in the tumor environment. Consequently, genetic ablation of St2 reduces Treg infiltration and concomitantly enhances the frequencies of effector CD8+ T cells, thereby restraining CRC. Mechanistically, IL-33 curtails IL-17 production by FOXP3+ Tregs and inhibits Th17 differentiation. In humans, numbers of activated ST2-expressing Tregs are increased in blood and tumor lesions of CRC patients, suggesting a similar mode of regulation. Together, these data indicate a central role of IL-33/ST2 signaling in shaping an immunosuppressive environment during intestinal tumorigenesis. Blockade of this pathway may provide a strategy to modulate the composition of CRC immune infiltrates.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Liew FY, Girard JP, Turnquist HR. Interleukin-33 in health and disease. Nat. Rev. Immunol. 2016;16:676–689. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

