Molecular Profiling of Inflammatory Bowel Disease: Is It Ready for Use in Clinical Decision-Making?
- PMID: 31167397
- PMCID: PMC6627070
- DOI: 10.3390/cells8060535
Molecular Profiling of Inflammatory Bowel Disease: Is It Ready for Use in Clinical Decision-Making?
Abstract
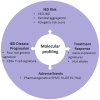
Inflammatory bowel disease (IBD) is a heterogeneous disorder in terms of age at onset, clinical phenotypes, severity, disease course, and response to therapy. This underlines the need for predictive and precision medicine that can optimize diagnosis and disease management, provide more cost-effective strategies, and minimize the risk of adverse events. Ideally, we can leverage molecular profiling to predict the risk to develop IBD and disease progression. Despite substantial successes of genome-wide association studies in the identification of genetic variants affecting IBD susceptibility, molecular profiling of disease onset and progression as well as of treatment responses has lagged behind. Still, thanks to technological advances and good study designs, predicting phenotypes using genomics and transcriptomics in IBD has been rapidly evolving. In this review, we summarize the current status of prediction of disease risk, clinical course, and response to therapy based on clinical case presentations. We also discuss the potential and limitations of the currently used approaches.
Keywords: genetics; inflammatory bowel disease; molecular profiling; transcriptomics.
Conflict of interest statement
H.-S. Lee and I. Cleynen declare no conflict of interest.
Figures


References
-
- Silverberg M.S., Satsangi J., Ahmad T., Arnott I.D., Bernstein C.N., Brant S.R., Caprilli R., Colombel J.F., Gasche C., Geboes K., et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: Report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can. J. Gastroenterol. 2005;19:5a–36a. doi: 10.1155/2005/269076. - DOI - PubMed
-
- Liu J.Z., van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A., Ripke S., Lee J.C., Jostins L., Shah T., et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47:979–986. doi: 10.1038/ng.3359. - DOI - PMC - PubMed
-
- de Lange K.M., Moutsianas L., Lee J.C., Lamb C.A., Luo Y., Kennedy N.A., Jostins L., Rice D.L., Gutierrez-Achury J., Ji S.G., et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 2017;49:256–261. doi: 10.1038/ng.3760. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

