Protein painting, an optimized MS-based technique, reveals functionally relevant interfaces of the PD-1/PD-L1 complex and the YAP2/ZO-1 complex
- PMID: 31167787
- PMCID: PMC6643031
- DOI: 10.1074/jbc.RA118.007310
Protein painting, an optimized MS-based technique, reveals functionally relevant interfaces of the PD-1/PD-L1 complex and the YAP2/ZO-1 complex
Abstract
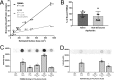
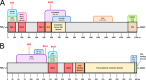
Protein-protein interactions lie at the heart of many biological processes and therefore represent promising drug targets. Despite this opportunity, identification of protein-protein interfaces remains challenging. We have previously developed a method that relies on coating protein surfaces with small-molecule dyes to discriminate between solvent-accessible protein surfaces and hidden interface regions. Dye-bound, solvent-accessible protein regions resist trypsin digestion, whereas hidden interface regions are revealed by denaturation and sequenced by MS. The small-molecule dyes bind promiscuously and with high affinity, but their binding mechanism is unknown. Here, we report on the optimization of a novel dye probe used in protein painting, Fast Blue B + naphthionic acid, and show that its affinity for proteins strongly depends on hydrophobic moieties that we call here "hydrophobic clamps." We demonstrate the utility of this probe by sequencing the protein-protein interaction regions between the Hippo pathway protein Yes-associated protein 2 (YAP2) and tight junction protein 1 (TJP1 or ZO-1), uncovering interactions via the known binding domain as well as ZO-1's MAGUK domain and YAP's N-terminal proline-rich domain. Additionally, we demonstrate how residues predicted by protein painting are present exclusively in the complex interface and how these residues may guide the development of peptide inhibitors using a case study of programmed cell death protein 1 (PD-1) and programmed cell death 1 ligand 1 (PD-L1). Inhibitors designed around the PD-1/PD-L1 interface regions identified via protein painting effectively disrupted complex formation, with the most potent inhibitor having an IC50 of 5 μm.
Keywords: PD-L1; Yes-associated protein (YAP); ZO-1; cell signaling; immune checkpoint inhibitor; mass spectrometry (MS); programmed cell death protein 1 (PD-1); protein painting; protein-protein interaction; structural biology.
© 2019 Haymond et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


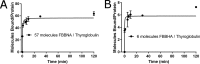

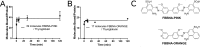

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

