The population history of northeastern Siberia since the Pleistocene
- PMID: 31168093
- PMCID: PMC7617447
- DOI: 10.1038/s41586-019-1279-z
The population history of northeastern Siberia since the Pleistocene
Abstract
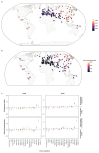
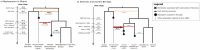
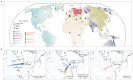
Northeastern Siberia has been inhabited by humans for more than 40,000 years but its deep population history remains poorly understood. Here we investigate the late Pleistocene population history of northeastern Siberia through analyses of 34 newly recovered ancient genomes that date to between 31,000 and 600 years ago. We document complex population dynamics during this period, including at least three major migration events: an initial peopling by a previously unknown Palaeolithic population of 'Ancient North Siberians' who are distantly related to early West Eurasian hunter-gatherers; the arrival of East Asian-related peoples, which gave rise to 'Ancient Palaeo-Siberians' who are closely related to contemporary communities from far-northeastern Siberia (such as the Koryaks), as well as Native Americans; and a Holocene migration of other East Asian-related peoples, who we name 'Neo-Siberians', and from whom many contemporary Siberians are descended. Each of these population expansions largely replaced the earlier inhabitants, and ultimately generated the mosaic genetic make-up of contemporary peoples who inhabit a vast area across northern Eurasia and the Americas.
Conflict of interest statement
Reprints and permissions information is available at
Figures








Comment in
-
The lineages of the first humans to reach northeastern Siberia and the Americas.Nature. 2019 Jun;570(7760):170-172. doi: 10.1038/d41586-019-01374-5. Nature. 2019. PMID: 31182830 No abstract available.
References
-
- Pitulko VV, et al. The Yana RHS Site: Humans in the Arctic Before the Last Glacial Maximum. Science. 2004;303:52–56. - PubMed
-
- Pitulko VV, Nikolskiy PA, Basilyan A, Pavlova EY. In: Paleoamerican Odyssey. Graf KE, Ketron CV, Waters MR, editors. Texas A&M University Press; 2014. Human Habitation in Arctic Western Beringia Prior to the LGM.
Methods references
-
- Allentoft ME, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522:167–172. - PubMed
-
- Orlando L, et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499:74. - PubMed
-
- Dabney J, Meyer M. Length and GC-biases during sequencing library amplification: a comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries. BioTechniques. 2012;52:87–94. - PubMed

