Modeling longitudinal imaging biomarkers with parametric Bayesian multi-task learning
- PMID: 31168892
- PMCID: PMC6679792
- DOI: 10.1002/hbm.24682
Modeling longitudinal imaging biomarkers with parametric Bayesian multi-task learning
Abstract
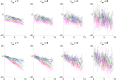
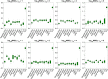
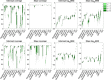
Longitudinal imaging biomarkers are invaluable for understanding the course of neurodegeneration, promising the ability to track disease progression and to detect disease earlier than cross-sectional biomarkers. To properly realize their potential, biomarker trajectory models must be robust to both under-sampling and measurement errors and should be able to integrate multi-modal information to improve trajectory inference and prediction. Here we present a parametric Bayesian multi-task learning based approach to modeling univariate trajectories across subjects that addresses these criteria. Our approach learns multiple subjects' trajectories within a single model that allows for different types of information sharing, that is, coupling, across subjects. It optimizes a combination of uncoupled, fully coupled and kernel coupled models. Kernel-based coupling allows linking subjects' trajectories based on one or more biomarker measures. We demonstrate this using Alzheimer's Disease Neuroimaging Initiative (ADNI) data, where we model longitudinal trajectories of MRI-derived cortical volumes in neurodegeneration, with coupling based on APOE genotype, cerebrospinal fluid (CSF) and amyloid PET-based biomarkers. In addition to detecting established disease effects, we detect disease related changes within the insula that have not received much attention within the literature. Due to its sensitivity in detecting disease effects, its competitive predictive performance and its ability to learn the optimal parameter covariance from data rather than choosing a specific set of random and fixed effects a priori, we propose that our model can be used in place of or in addition to linear mixed effects models when modeling biomarker trajectories. A software implementation of the method is publicly available.
Keywords: Alzheimer's disease; Bayesian analysis; biomarkers; longitudinal analysis; machine learning; multimodal analysis; structural MRI.
© 2019 The Authors. Human Brain Mapping published by Wiley Periodicals, Inc.
Figures



References
-
- Abadi, M. , Barham, P. , Chen, J. , Chen, Z. , Davis, A. , Dean, J. , Devin, M. , Ghemawat, S. , Irving, G. , Isard, M. , Kudlur, M. , Levenberg, J. , Monga, R. , Moore, S. , Murray, D.G. , Steiner, B. , Tucker, P. , Vasudevan, V. , Warden, P. , Wicke, M. , Yu, Y. , Zheng, X. , 2016. TensorFlow: A system for large‐scale machine learning.
-
- Bernal‐Rusiel, J. L. , Greve, D. N. , Reuter, M. , Fischl, B. , Sabuncu, M. R. , & Alzheimer's Disease Neuroimaging Initiative . (2013). Statistical analysis of longitudinal neuroimage data with linear mixed effects models. NeuroImage, 66, 249–260. 10.1016/j.neuroimage.2012.10.065 - DOI - PMC - PubMed
-
- Bishop, C. M. (2007). Pattern recognition and machine learning. New York: Springer.
Publication types
MeSH terms
Substances
Grants and funding
- FP7-ICT-2011-9-601055/FP7 Information and Communication Technologies/International
- BW.mn.BRC10269/National Institute for Health Research/International
- MR/J01107X/1/MRC_/Medical Research Council/United Kingdom
- M006093/Engineering and Physical Sciences Research Council/International
- EP/K005278/Engineering and Physical Sciences Research Council/International
- 666992/Horizon 2020 Framework Programme/International
- EP/L016478/1/Engineering and Physical Sciences Research Council/International
- U01 AG024904/AG/NIA NIH HHS/United States
- EP/J020990/1/Engineering and Physical Sciences Research Council/International
- MR/L016311/1/MRC_/Medical Research Council/United Kingdom
- J020990/Engineering and Physical Sciences Research Council/International
- 91716415/Nederlandse Organisatie voor Wetenschappelijk Onderzoek/International
- U19 AG024904/AG/NIA NIH HHS/United States
- M020533/Engineering and Physical Sciences Research Council/International
- EP/H046410/1/Engineering and Physical Sciences Research Council/International
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

