Epigenetics recording varied environment and complex cell events represents the origin of cellular aging
- PMID: 31168969
- PMCID: PMC6586999
- DOI: 10.1631/jzus.B1800507
Epigenetics recording varied environment and complex cell events represents the origin of cellular aging
Abstract
Although a relationship between epigenetics and aging phenotypic changes has been established, a theoretical explanation of the intrinsic connection between the epigenetics and aging is lacking. In this essay, we propose that epigenetic recording of varied cell environment and complex history could be an origin of cellular aging. Through epigenetic modifications, the environment and historical events can induce the chromatin template into an activated or repressive accessible structure, thereby shaping the DNA template into a spectrum of chromatin states. The inner nature of diversity and conflicts born by the cell environment and its historical events are hence recorded into the chromatin template. This could result in a dissipated spectrum of the chromatin state and chaos in overall gene expression. An unavoidable degradation of epigenome entropy, similar to Shannon entropy, would be consequently induced. The resultant disorder in epigenome, characterized by corrosion of epigenome entropy as reflected in chromatin template, can be stably memorized and propagated through cell division. Furthermore, the hysteretic nature of epigenetics responding to the emerging environment could exacerbate the degradation of epigenome entropy. As well as stochastic errors, we propose that outside entropy (or chaos) derived from the varied environment and complex cell history, gradually input and imprinted into the chromatin via epigenetic modifications, would lead inevitably to cellular aging, the extent of which could be aggravated by hysteresis of epigenetics without error erasing and correction.
Keywords: Epigenetics; Environment; Cell event; Cellular aging; Epigenome entropy; DNA methylation.
Conflict of interest statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Figures

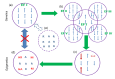


References
-
- Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell, 5th Ed. Garland Science Taylor and Francis Group, New York; 2008. pp. 411–477.
-
- Allis CD, Caparros ML, Jenuwein T, et al. Overview and concepts. In: Allis CD Caparros ML, Jenuwein T et al., editors. Epigenetics, 2nd Ed. Cold Spring Harbor, New York; 2015. pp. 47–115.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

