A mutation in the endonuclease domain of mouse MLH3 reveals novel roles for MutLγ during crossover formation in meiotic prophase I
- PMID: 31170160
- PMCID: PMC6588253
- DOI: 10.1371/journal.pgen.1008177
A mutation in the endonuclease domain of mouse MLH3 reveals novel roles for MutLγ during crossover formation in meiotic prophase I
Abstract
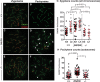
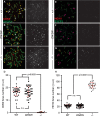
During meiotic prophase I, double-strand breaks (DSBs) initiate homologous recombination leading to non-crossovers (NCOs) and crossovers (COs). In mouse, 10% of DSBs are designated to become COs, primarily through a pathway dependent on the MLH1-MLH3 heterodimer (MutLγ). Mlh3 contains an endonuclease domain that is critical for resolving COs in yeast. We generated a mouse (Mlh3DN/DN) harboring a mutation within this conserved domain that is predicted to generate a protein that is catalytically inert. Mlh3DN/DN males, like fully null Mlh3-/- males, have no spermatozoa and are infertile, yet spermatocytes have grossly normal DSBs and synapsis events in early prophase I. Unlike Mlh3-/- males, mutation of the endonuclease domain within MLH3 permits normal loading and frequency of MutLγ in pachynema. However, key DSB repair factors (RAD51) and mediators of CO pathway choice (BLM helicase) persist into pachynema in Mlh3DN/DN males, indicating a temporal delay in repair events and revealing a mechanism by which alternative DSB repair pathways may be selected. While Mlh3DN/DN spermatocytes retain only 22% of wildtype chiasmata counts, this frequency is greater than observed in Mlh3-/- males (10%), suggesting that the allele may permit partial endonuclease activity, or that other pathways can generate COs from these MutLγ-defined repair intermediates in Mlh3DN/DN males. Double mutant mice homozygous for the Mlh3DN/DN and Mus81-/- mutations show losses in chiasmata close to those observed in Mlh3-/- males, indicating that the MUS81-EME1-regulated crossover pathway can only partially account for the increased residual chiasmata in Mlh3DN/DN spermatocytes. Our data demonstrate that mouse spermatocytes bearing the MLH1-MLH3DN/DN complex display the proper loading of factors essential for CO resolution (MutSγ, CDK2, HEI10, MutLγ). Despite these functions, mice bearing the Mlh3DN/DN allele show defects in the repair of meiotic recombination intermediates and a loss of most chiasmata.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Allers T, Lichten M (2001) Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol Cell 8: 225–231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

