MTR-Viewer: identifying regions within genes under purifying selection
- PMID: 31170280
- PMCID: PMC6602522
- DOI: 10.1093/nar/gkz457
MTR-Viewer: identifying regions within genes under purifying selection
Abstract
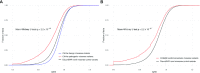
Advances in genomic sequencing have enormous potential to revolutionize personalized medicine, however distinguishing disease-causing from benign variants remains a challenge. The increasing number of human genome and exome sequences available has revealed areas where unfavourable variation is removed through purifying selection. Here, we present the MTR-Viewer, a web-server enabling easy visualization at the gene or variant level of the Missense Tolerance Ratio (MTR), a measure of regional intolerance to missense variation calculated using variation from 240 000 exome and genome sequences. The MTR-Viewer enables exploration of MTR calculations, using different sliding windows, for over 18 000 human protein-coding genes and 85 000 alternative transcripts. Users can also view MTR scores calculated for specific ethnicities, to enable easy exploration of regions that may be under different selective pressure. The spatial distribution of population and known disease variants is also displayed on the protein's domain structure. Intolerant regions were found to be highly enriched for ClinVar pathogenic and COSMIC somatic missense variants (Mann-Whitney U test P < 2.2 × 10-16). As the MTR is not biased by known domains and protein features, it can highlight functionally important regions within genes overlooked or inaccessible by traditional methods. MTR-Viewer is freely available via a user friendly web-server at http://biosig.unimelb.edu.au/mtr-viewer/.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

