Genome-Wide Association Study and Cost-Efficient Genomic Predictions for Growth and Fillet Yield in Nile Tilapia (Oreochromis niloticus)
- PMID: 31171566
- PMCID: PMC6686944
- DOI: 10.1534/g3.119.400116
Genome-Wide Association Study and Cost-Efficient Genomic Predictions for Growth and Fillet Yield in Nile Tilapia (Oreochromis niloticus)
Abstract
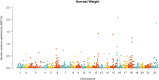
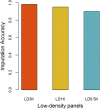
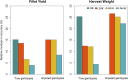
Fillet yield (FY) and harvest weight (HW) are economically important traits in Nile tilapia production. Genetic improvement of these traits, especially for FY, are lacking, due to the absence of efficient methods to measure the traits without sacrificing fish and the use of information from relatives to selection. However, genomic information could be used by genomic selection to improve traits that are difficult to measure directly in selection candidates, as in the case of FY. The objectives of this study were: (i) to perform genome-wide association studies (GWAS) to dissect the genetic architecture of FY and HW, (ii) to evaluate the accuracy of genotype imputation and (iii) to assess the accuracy of genomic selection using true and imputed low-density (LD) single nucleotide polymorphism (SNP) panels to determine a cost-effective strategy for practical implementation of genomic information in tilapia breeding programs. The data set consisted of 5,866 phenotyped animals and 1,238 genotyped animals (108 parents and 1,130 offspring) using a 50K SNP panel. The GWAS were performed using all genotyped and phenotyped animals. The genotyped imputation was performed from LD panels (LD0.5K, LD1K and LD3K) to high-density panel (HD), using information from parents and 20% of offspring in the reference set and the remaining 80% in the validation set. In addition, we tested the accuracy of genomic selection using true and imputed genotypes comparing the accuracy obtained from pedigree-based best linear unbiased prediction (PBLUP) and genomic predictions. The results from GWAS supports evidence of the polygenic nature of FY and HW. The accuracy of imputation ranged from 0.90 to 0.98 for LD0.5K and LD3K, respectively. The accuracy of genomic prediction outperformed the estimated breeding value from PBLUP. The use of imputation for genomic selection resulted in an increased relative accuracy independent of the trait and LD panel analyzed. The present results suggest that genotype imputation could be a cost-effective strategy for genomic selection in Nile tilapia breeding programs.
Keywords: GWAS; GenPred; Oreochromis niloticus; Shared Data Resources; complex traits; cost-efficient; genomic prediction; genotype imputation; low-density panel.
Copyright © 2019 Yoshida et al.
Figures

Similar articles
-
Genomic selection models substantially improve the accuracy of genetic merit predictions for fillet yield and body weight in rainbow trout using a multi-trait model and multi-generation progeny testing.Genet Sel Evol. 2023 Feb 9;55(1):11. doi: 10.1186/s12711-023-00782-6. Genet Sel Evol. 2023. PMID: 36759760 Free PMC article.
-
Assets of imputation to ultra-high density for productive and functional traits.J Dairy Sci. 2013 Sep;96(9):6047-58. doi: 10.3168/jds.2013-6793. Epub 2013 Jun 28. J Dairy Sci. 2013. PMID: 23810591
-
Genotype Imputation To Improve the Cost-Efficiency of Genomic Selection in Farmed Atlantic Salmon.G3 (Bethesda). 2017 Apr 3;7(4):1377-1383. doi: 10.1534/g3.117.040717. G3 (Bethesda). 2017. PMID: 28250015 Free PMC article.
-
Evaluation of measures of correctness of genotype imputation in the context of genomic prediction: a review of livestock applications.Animal. 2014 Nov;8(11):1743-53. doi: 10.1017/S1751731114001803. Epub 2014 Jul 21. Animal. 2014. PMID: 25045914 Review.
-
Sunflower Hybrid Breeding: From Markers to Genomic Selection.Front Plant Sci. 2018 Jan 17;8:2238. doi: 10.3389/fpls.2017.02238. eCollection 2017. Front Plant Sci. 2018. PMID: 29387071 Free PMC article. Review.
Cited by
-
Evaluation of low-density SNP panels and imputation for cost-effective genomic selection in four aquaculture species.Front Genet. 2023 May 11;14:1194266. doi: 10.3389/fgene.2023.1194266. eCollection 2023. Front Genet. 2023. PMID: 37252666 Free PMC article.
-
Fine-tuning GBS data with comparison of reference and mock genome approaches for advancing genomic selection in less studied farmed species.BMC Genomics. 2025 Feb 5;26(1):111. doi: 10.1186/s12864-025-11296-4. BMC Genomics. 2025. PMID: 39910437 Free PMC article.
-
Genome-Wide Patterns of Population Structure and Linkage Disequilibrium in Farmed Nile Tilapia (Oreochromis niloticus).Front Genet. 2019 Sep 4;10:745. doi: 10.3389/fgene.2019.00745. eCollection 2019. Front Genet. 2019. PMID: 31552083 Free PMC article.
-
Genomic Selection and Genome-wide Association Study for Feed-Efficiency Traits in a Farmed Nile Tilapia (Oreochromis niloticus) Population.Front Genet. 2021 Sep 20;12:737906. doi: 10.3389/fgene.2021.737906. eCollection 2021. Front Genet. 2021. PMID: 34616434 Free PMC article.
-
Optimizing Low-Cost Genotyping and Imputation Strategies for Genomic Selection in Atlantic Salmon.G3 (Bethesda). 2020 Feb 6;10(2):581-590. doi: 10.1534/g3.119.400800. G3 (Bethesda). 2020. PMID: 31826882 Free PMC article.
References
-
- Barria A., Christensen K. A., Yoshida G. M., Correa K., Jedlicki A. et al. , 2018. Genomic predictions and genome-wide association study of resistance against Piscirickettsia salmonis in coho salmon (Oncorhynchus kisutch) using ddRAD sequencing. G3 Genes, Genomes. Genet. 8: 1183–1194. 10.1534/g3.118.200053 - DOI - PMC - PubMed
-
- Barría A., Christensen K. A., Yoshida G., Jedlicki A., Leong J. S. et al. , 2019. Whole genome linkage disequilibrium and effective population size in a coho salmon (Oncorhynchus kisutch) breeding population using a high density SNP array. Front. Genet. 10: 498 10.3389/fgene.2019.00498 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
