Phosphorylation of G3BP1-S149 does not influence stress granule assembly
- PMID: 31171631
- PMCID: PMC6605800
- DOI: 10.1083/jcb.201801214
Phosphorylation of G3BP1-S149 does not influence stress granule assembly
Abstract
Tourrière et al. (2013. J. Cell Biol. https://doi.org/10.1083/jcb.200212128) reported that G3BP1-S149 dephosphorylation promotes stress granule formation. We show that constructs used to establish this conclusion contain additional mutations causing these phenotypes, and that S149 phosphorylation status does not change upon stress.
© 2019 Panas et al.
Figures
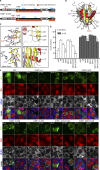
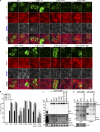

Comment in
-
Reply to "Phosphorylation of G3BP1-S149 does not influence stress granule assembly".J Cell Biol. 2019 Jul 1;218(7):2433-2434. doi: 10.1083/jcb.201905105. Epub 2019 Jun 6. J Cell Biol. 2019. PMID: 31171633 Free PMC article.
Comment on
-
The RasGAP-associated endoribonuclease G3BP assembles stress granules.J Cell Biol. 2003 Mar 17;160(6):823-31. doi: 10.1083/jcb.200212128. J Cell Biol. 2003. Retraction in: J Cell Biol. 2023 Nov 6;222(11):e20021212808022023r. doi: 10.1083/jcb.20021212808022023r. PMID: 12642610 Free PMC article. Retracted.
References
-
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., and Terwilliger T.C.. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954. 10.1107/S0907444902016657 - DOI - PubMed
-
- Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B. III, Snoeyink J., Richardson J.S., and Richardson D.C.. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35:W375–W383 10.1093/nar/gkm216 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

