Illuminating the Onco-GPCRome: Novel G protein-coupled receptor-driven oncocrine networks and targets for cancer immunotherapy
- PMID: 31171722
- PMCID: PMC6643028
- DOI: 10.1074/jbc.REV119.005601
Illuminating the Onco-GPCRome: Novel G protein-coupled receptor-driven oncocrine networks and targets for cancer immunotherapy
Abstract
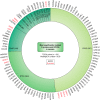
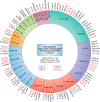
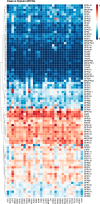
G protein-coupled receptors (GPCRs) are the largest gene family of cell membrane-associated molecules mediating signal transmission, and their involvement in key physiological functions is well-established. The ability of GPCRs to regulate a vast array of fundamental biological processes, such as cardiovascular functions, immune responses, hormone and enzyme release from endocrine and exocrine glands, neurotransmission, and sensory perception (e.g. vision, odor, and taste), is largely due to the diversity of these receptors and the layers of their downstream signaling circuits. Dysregulated expression and aberrant functions of GPCRs have been linked to some of the most prevalent human diseases, which renders GPCRs one of the top targets for pharmaceutical drug development. However, the study of the role of GPCRs in tumor biology has only just begun to make headway. Recent studies have shown that GPCRs can contribute to the many facets of tumorigenesis, including proliferation, survival, angiogenesis, invasion, metastasis, therapy resistance, and immune evasion. Indeed, GPCRs are widely dysregulated in cancer and yet are underexploited in oncology. We present here a comprehensive analysis of GPCR gene expression, copy number variation, and mutational signatures in 33 cancer types. We also highlight the emerging role of GPCRs as part of oncocrine networks promoting tumor growth, dissemination, and immune evasion, and we stress the potential benefits of targeting GPCRs and their signaling circuits in the new era of precision medicine and cancer immunotherapies.
Keywords: G protein; G protein–coupled receptor (GPCR); cancer; drug repurposing; immunotherapy; oncocrine signaling; precision therapies; signal transduction.
© 2019 Wu et al.
Conflict of interest statement
J. S. G. is a member of the Scientific Advisory Board of Oncoceutics Inc. and Domain Therapeutics
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R01 GM074024/GM/NIGMS NIH HHS/United States
- R33 CA225291/CA/NCI NIH HHS/United States
- U24 CA194107/CA/NCI NIH HHS/United States
- U24 CA220341/CA/NCI NIH HHS/United States
- P30 CA023100/CA/NCI NIH HHS/United States
- T32 CA067754/CA/NCI NIH HHS/United States
- R01 CA172513/CA/NCI NIH HHS/United States
- U01 CA217885/CA/NCI NIH HHS/United States
- U01 CA196406/CA/NCI NIH HHS/United States
- U24 CA248457/CA/NCI NIH HHS/United States
- R01 HG009285/HG/NHGRI NIH HHS/United States
- U01 DE028227/DE/NIDCR NIH HHS/United States
- U54 CA209891/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

