Genomic and Metagenomic Approaches for Predictive Surveillance of Emerging Pathogens and Antibiotic Resistance
- PMID: 31172511
- PMCID: PMC6692204
- DOI: 10.1002/cpt.1535
Genomic and Metagenomic Approaches for Predictive Surveillance of Emerging Pathogens and Antibiotic Resistance
Abstract
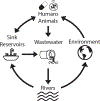
Antibiotic-resistant organisms (AROs) are a major concern to public health worldwide. While antibiotics have been naturally produced by environmental bacteria for millions of years, modern widespread use of antibiotics has enriched resistance mechanisms in human-impacted bacterial environments. Antibiotic resistance genes (ARGs) continue to emerge and spread rapidly. To combat the global threat of antibiotic resistance, researchers must develop methods to rapidly characterize AROs and ARGs, monitor their spread across space and time, and identify novel ARGs and resistance pathways. We review how high-throughput sequencing-based methods can be combined with classic culture-based assays to characterize, monitor, and track AROs and ARGs. Then, we evaluate genomic and metagenomic methods for identifying ARGs and biosynthetic pathways for novel antibiotics from genomic data sets. Together, these genomic analyses can improve surveillance and prediction of emerging resistance threats and accelerate the development of new antibiotic therapies to combat resistance.
© 2019 The Authors Clinical Pharmacology & Therapeutics © 2019 American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
Conflict of Interest: The authors declared no competing interests for this work.
Figures
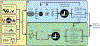

References
-
- Brown ED & Wright GD Antibacterial drug discovery in the resistance era. Nature 529, 336–43 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

