Changing substrate specificity and iteration of amino acid chain elongation in glucosinolate biosynthesis through targeted mutagenesis of Arabidopsis methylthioalkylmalate synthase 1
- PMID: 31175145
- PMCID: PMC6603273
- DOI: 10.1042/BSR20190446
Changing substrate specificity and iteration of amino acid chain elongation in glucosinolate biosynthesis through targeted mutagenesis of Arabidopsis methylthioalkylmalate synthase 1
Abstract
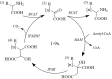
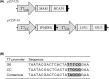
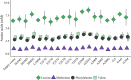
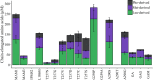
Methylthioalkylmalate synthases catalyse the committing step of amino acid chain elongation in glucosinolate biosynthesis. As such, this group of enzymes plays an important role in determining the glucosinolate composition of Brassicaceae species, including Arabidopsis thaliana Based on protein structure modelling of MAM1 from A. thaliana and analysis of 57 MAM sequences from Brassicaceae species, we identified four polymorphic residues likely to interact with the 2-oxo acid substrate. Through site-directed mutagenesis, the natural variation in these residues and the effect on product composition were investigated. Fifteen MAM1 variants as well as the native MAM1 and MAM3 from A. thaliana were characterised by heterologous expression of the glucosinolate chain elongation pathway in Escherichia coli Detected products derived from leucine, methionine or phenylalanine were elongated with up to six methylene groups. Product profile and accumulation were changed in 14 of the variants, demonstrating the relevance of the identified residues. The majority of the single amino acid substitutions decreased the length of methionine-derived products, while approximately half of the substitutions increased the phenylalanine-derived products. Combining two substitutions enabled the MAM1 variant to increase the number of elongation rounds of methionine from three to four. Notably, characterisation of the native MAMs indicated that MAM1 and not MAM3 is responsible for homophenylalanine production. This hypothesis was confirmed by glucosinolate analysis in mam1 and mam3 mutants of A. thaliana.
Keywords: Escherichia coli; biotechnology; enzyme activity; structural biology.
© 2019 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures



Comment in
-
Glucosinolate biosynthesis: role of MAM synthase and its perspectives.Biosci Rep. 2021 Oct 29;41(10):BSR20211634. doi: 10.1042/BSR20211634. Biosci Rep. 2021. PMID: 34545928 Free PMC article.
Similar articles
-
Expression pattern of the glucosinolate side chain biosynthetic genes MAM1 and MAM3 of Arabidopsis thaliana in different organs and developmental stages.Plant Physiol Biochem. 2012 Apr;53:77-83. doi: 10.1016/j.plaphy.2012.01.015. Epub 2012 Jan 28. Plant Physiol Biochem. 2012. PMID: 22336876
-
MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis.Plant Physiol. 2007 May;144(1):60-71. doi: 10.1104/pp.106.091579. Epub 2007 Mar 16. Plant Physiol. 2007. PMID: 17369439 Free PMC article.
-
From amino acid to glucosinolate biosynthesis: protein sequence changes in the evolution of methylthioalkylmalate synthase in Arabidopsis.Plant Cell. 2011 Jan;23(1):38-53. doi: 10.1105/tpc.110.079269. Epub 2011 Jan 4. Plant Cell. 2011. PMID: 21205930 Free PMC article.
-
Structural Studies of Aliphatic Glucosinolate Chain-Elongation Enzymes.Antioxidants (Basel). 2021 Sep 21;10(9):1500. doi: 10.3390/antiox10091500. Antioxidants (Basel). 2021. PMID: 34573132 Free PMC article. Review.
-
Understanding of MYB Transcription Factors Involved in Glucosinolate Biosynthesis in Brassicaceae.Molecules. 2017 Sep 14;22(9):1549. doi: 10.3390/molecules22091549. Molecules. 2017. PMID: 28906468 Free PMC article. Review.
Cited by
-
Developing multifunctional crops by engineering Brassicaceae glucosinolate pathways.Plant Commun. 2023 Jul 10;4(4):100565. doi: 10.1016/j.xplc.2023.100565. Epub 2023 Feb 23. Plant Commun. 2023. PMID: 36823985 Free PMC article. Review.
-
Genomic Origin and Diversification of the Glucosinolate MAM Locus.Front Plant Sci. 2020 Jun 4;11:711. doi: 10.3389/fpls.2020.00711. eCollection 2020. Front Plant Sci. 2020. PMID: 32582245 Free PMC article.
-
Characterization of HphA: The First Enzyme in the Homologation Pathway of l-Phenylalanine and l-Tyrosine.Chembiochem. 2024 Aug 19;25(16):e202400369. doi: 10.1002/cbic.202400369. Epub 2024 Aug 2. Chembiochem. 2024. PMID: 38896437 Free PMC article.
-
Effects of Plant Hormones, Metal Ions, Salinity, Sugar, and Chemicals Pollution on Glucosinolate Biosynthesis in Cruciferous Plant.Front Plant Sci. 2022 Apr 28;13:856442. doi: 10.3389/fpls.2022.856442. eCollection 2022. Front Plant Sci. 2022. PMID: 35574082 Free PMC article. Review.
-
Co-Regulation of Clustered and Neo-Functionalized Genes in Plant-Specialized Metabolism.Plants (Basel). 2020 May 13;9(5):622. doi: 10.3390/plants9050622. Plants (Basel). 2020. PMID: 32414181 Free PMC article. Review.
References
-
- Benderoth M., Pfalz M. and Kroymann J. (2009) Methylthioalkylmalate synthases: genetics, ecology and evolution. Phytochem. Rev. 8, 255–26810.1007/s11101-008-9097-1 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

