AtTRAPPC11/ROG2: A Role for TRAPPs in Maintenance of the Plant Trans-Golgi Network/Early Endosome Organization and Function
- PMID: 31175171
- PMCID: PMC6713296
- DOI: 10.1105/tpc.19.00110
AtTRAPPC11/ROG2: A Role for TRAPPs in Maintenance of the Plant Trans-Golgi Network/Early Endosome Organization and Function
Abstract
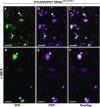
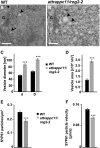
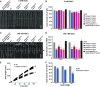
The dynamic trans-Golgi network/early endosome (TGN/EE) facilitates cargo sorting and trafficking and plays a vital role in plant development and environmental response. Transport protein particles (TRAPPs) are multi-protein complexes acting as guanine nucleotide exchange factors and possibly as tethers, regulating intracellular trafficking. TRAPPs are essential in all eukaryotic cells and are implicated in a number of human diseases. It has been proposed that they also play crucial roles in plants; however, our current knowledge about the structure and function of plant TRAPPs is very limited. Here, we identified and characterized AtTRAPPC11/RESPONSE TO OLIGOGALACTURONIDE2 (AtTRAPPC11/ROG2), a TGN/EE-associated, evolutionarily conserved TRAPP protein in Arabidopsis (Arabidopsis thaliana). AtTRAPPC11/ROG2 regulates TGN integrity, as evidenced by altered TGN/EE association of several residents, including SYNTAXIN OF PLANTS61, and altered vesicle morphology in attrappc11/rog2 mutants. Furthermore, endocytic traffic and brefeldin A body formation are perturbed in attrappc11/rog2, suggesting a role for AtTRAPPC11/ROG2 in regulation of endosomal function. Proteomic analysis showed that AtTRAPPC11/ROG2 defines a hitherto uncharacterized TRAPPIII complex in plants. In addition, attrappc11/rog2 mutants are hypersensitive to salinity, indicating an undescribed role of TRAPPs in stress responses. Overall, our study illustrates the plasticity of the endomembrane system through TRAPP protein functions and opens new avenues to explore this dynamic network.
© 2019 American Society of Plant Biologists. All rights reserved.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

