Tau local structure shields an amyloid-forming motif and controls aggregation propensity
- PMID: 31175300
- PMCID: PMC6555816
- DOI: 10.1038/s41467-019-10355-1
Tau local structure shields an amyloid-forming motif and controls aggregation propensity
Abstract
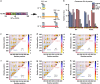
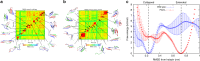
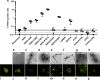
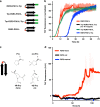
Tauopathies are neurodegenerative diseases characterized by intracellular amyloid deposits of tau protein. Missense mutations in the tau gene (MAPT) correlate with aggregation propensity and cause dominantly inherited tauopathies, but their biophysical mechanism driving amyloid formation is poorly understood. Many disease-associated mutations localize within tau's repeat domain at inter-repeat interfaces proximal to amyloidogenic sequences, such as 306VQIVYK311. We use cross-linking mass spectrometry, recombinant protein and synthetic peptide systems, in silico modeling, and cell models to conclude that the aggregation-prone 306VQIVYK311 motif forms metastable compact structures with its upstream sequence that modulates aggregation propensity. We report that disease-associated mutations, isomerization of a critical proline, or alternative splicing are all sufficient to destabilize this local structure and trigger spontaneous aggregation. These findings provide a biophysical framework to explain the basis of early conformational changes that may underlie genetic and sporadic tau pathogenesis.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

