PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis
- PMID: 31177901
- PMCID: PMC6999623
- DOI: 10.1080/15548627.2019.1628520
PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis
Abstract
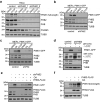
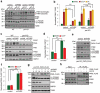
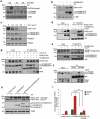
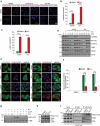
Mitophagy, which is a conserved cellular process for selectively removing damaged or unwanted mitochondria, is critical for mitochondrial quality control and the maintenance of normal cellular physiology. However, the precise mechanisms underlying mitophagy remain largely unknown. Prior studies on mitophagy focused on the events in the mitochondrial outer membrane. PHB2 (prohibitin 2), which is a highly conserved membrane scaffold protein, was recently identified as a novel inner membrane mitophagy receptor that mediates mitophagy. Here, we report a new signaling pathway for PHB2-mediated mitophagy. Upon mitochondrial membrane depolarization or misfolded protein aggregation, PHB2 depletion destabilizes PINK1 in the mitochondria, which blocks the mitochondrial recruitment of PRKN/Parkin, ubiquitin and OPTN (optineurin), leading to an inhibition of mitophagy. In addition, PHB2 overexpression directly induces PRKN recruitment to the mitochondria. Moreover, PHB2-mediated mitophagy is dependent on the mitochondrial inner membrane protease PARL, which interacts with PHB2 and is activated upon PHB2 depletion. Furthermore, PGAM5, which is processed by PARL, participates in PHB2-mediated PINK1 stabilization. Finally, a ligand of PHB proteins that we synthesized, called FL3, was found to strongly inhibit PHB2-mediated mitophagy and to effectively block cancer cell growth and energy production at nanomolar concentrations. Thus, our findings reveal that the PHB2-PARL-PGAM5-PINK1 axis is a novel pathway of PHB2-mediated mitophagy and that targeting PHB2 with the chemical compound FL3 is a promising strategy for cancer therapy.Abbreviations: AIFM1: apoptosis inducing factor mitochondria associated 1; ATP5F1A/ATP5A1: ATP synthase F1 subunit alpha; BAF: bafilomycin A1; CALCOCO2/NDP52: calcium binding and coiled-coil domain 2; CCCP: chemical reagent carbonyl cyanide m-chlorophenyl hydrazine; FL3: flavaglines compound 3; HSPD1/HSP60: heat shock protein family D (Hsp60) member 1; LC3B/MAP1LC3B: microtubule associated protein 1 light chain 3 beta; MEF: mouse embryo fibroblasts; MPP: mitochondrial-processing peptidase; MT-CO2/COX2: mitochondrially encoded cytochrome c oxidase II; MTS: mitochondrial targeting sequence; OA: oligomycin and antimycin A; OPTN: optineurin; OTC: ornithine carbamoyltransferase; PARL: presenilin associated rhomboid like; PBS: phosphate-buffered saline; PGAM5: PGAM family member 5, mitochondrial serine/threonine protein phosphatase; PHB: prohibitin; PHB2: prohibitin 2; PINK1: PTEN induced kinase 1; PRKN/Parkin: parkin RBR E3 ubiquitin protein ligase; Roc-A: rocaglamide A; TOMM20: translocase of outer mitochondrial membrane 20; TUBB: tubulin beta class I.
Keywords: Mitophagy; PARL; PGAM5; PHB2; PINK1-PRKN.
Figures



References
-
- Georgakopoulos ND, Wells G, Campanella M.. The pharmacological regulation of cellular mitophagy. Nat Chem Biol. 2017. January 19;13(2):136–146. PubMed PMID: 28103219. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
