Total copy number variation as a prognostic factor in adult astrocytoma subtypes
- PMID: 31177992
- PMCID: PMC6556960
- DOI: 10.1186/s40478-019-0746-y
Total copy number variation as a prognostic factor in adult astrocytoma subtypes
Erratum in
-
Publisher Correction to: Acta Neuropathologica Communications, volume 7.Acta Neuropathol Commun. 2019 Aug 14;7(1):131. doi: 10.1186/s40478-019-0784-5. Acta Neuropathol Commun. 2019. PMID: 31412936 Free PMC article.
Abstract
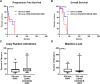
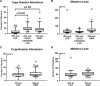
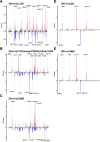
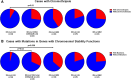
Since the discovery that IDH1/2 mutations confer a significantly better prognosis in astrocytomas, much work has been done to identify other molecular signatures to help further stratify lower-grade astrocytomas and glioblastomas, with the goal of accurately predicting clinical outcome and identifying potentially targetable mutations. In the present study, we subclassify 135 astrocytomas (67 IDH-wildtype and 68 IDH-mutant) from The Cancer Genome Atlas dataset (TCGA) on the basis of grade, IDH-status, and the previously established prognostic factors, CDK4 amplification and CDKN2A/B deletion, within the IDH-mutant groups. We analyzed these groups for total copy number variation (CNV), total mutation burden, chromothripsis, specific mutations, and amplifications/deletions of specific genes/chromosomal regions. Herein, we demonstrate that across all of these tumor groups, total CNV level is a relatively consistent prognostic factor. We also identified a trend towards increased levels of chromothripsis in tumors with lower progression-free survival (PFS) and overall survival (OS) intervals. While no significant differences were identified in overall mutation load, we did identify a significantly higher number of cases with mutations in genes with functions related to maintaining genomic stability in groups with higher mean CNV and worse PFS and OS intervals, particularly in the IDH-mutant groups. Our data further support the case for total CNV level as a potential prognostic factor in astrocytomas, and suggest mutations in genes responsible for overall genomic instability as a possible underlying mechanism for some astrocytomas with poor clinical outcome.
Keywords: Astrocytoma; CNV; Copy number variation; GBM; Glioblastoma; Glioma; TCGA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. - DOI - PMC - PubMed
-
- Brat DJ, Aldape K, Colman H, Holland EC, Louis DN, Jenkins RB, Kleinschmidt-DeMasters BK, Perry A, Reifenberger G, Stupp R, et al. cIMPACT-NOW update 3: recommended diagnostic criteria for “diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol. 2018;136:805–810. doi: 10.1007/s00401-018-1913-0. - DOI - PMC - PubMed
-
- Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

