Multi-Color Single-Molecule Imaging Uncovers Extensive Heterogeneity in mRNA Decoding
- PMID: 31178119
- PMCID: PMC6630898
- DOI: 10.1016/j.cell.2019.05.001
Multi-Color Single-Molecule Imaging Uncovers Extensive Heterogeneity in mRNA Decoding
Abstract
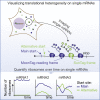
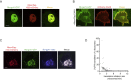
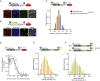
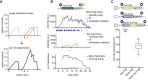
mRNA translation is a key step in decoding genetic information. Genetic decoding is surprisingly heterogeneous because multiple distinct polypeptides can be synthesized from a single mRNA sequence. To study translational heterogeneity, we developed the MoonTag, a fluorescence labeling system to visualize translation of single mRNAs. When combined with the orthogonal SunTag system, the MoonTag enables dual readouts of translation, greatly expanding the possibilities to interrogate complex translational heterogeneity. By placing MoonTag and SunTag sequences in different translation reading frames, each driven by distinct translation start sites, start site selection of individual ribosomes can be visualized in real time. We find that start site selection is largely stochastic but that the probability of using a particular start site differs among mRNA molecules and can be dynamically regulated over time. This study provides key insights into translation start site selection heterogeneity and provides a powerful toolbox to visualize complex translation dynamics.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures



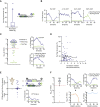
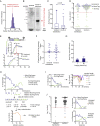

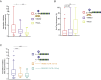

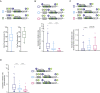
References
-
- Aitken C.E., Lorsch J.R. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2012;19:568–576. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

