A Genomic, Evolutionary, and Mechanistic Study of MCR-5 Action Suggests Functional Unification across the MCR Family of Colistin Resistance
- PMID: 31179217
- PMCID: PMC6548960
- DOI: 10.1002/advs.201900034
A Genomic, Evolutionary, and Mechanistic Study of MCR-5 Action Suggests Functional Unification across the MCR Family of Colistin Resistance
Abstract
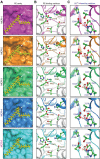
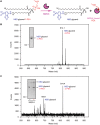
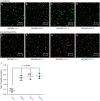
A growing number of mobile colistin resistance (MCR) proteins is threatening the renewed interest of colistin as a "last-resort" defense against carbapenem-resistant pathogens. Here, the comparative genomics of a large plasmid harboring mcr-5 from Aeromonas hydrophila and the structural/functional perspectives of MCR-5 action are reported. Whole genome sequencing has identified the loss of certain parts of the Tn3-type transposon typically associated with mcr-5, providing a clue toward its mobilization. Phylogeny of MCR-5 suggests that it is distinct from the MCR-1/2 sub-lineage, but might share a common ancestor of MCR-3/4. Domain-swapping analysis of MCR-5 elucidates that its two structural motifs (transmembrane domain and catalytic domain) are incompatible with its counterparts in MCR-1/2. Like the rest of the MCR family, MCR-5 exhibits a series of conservative features, including zinc-dependent active sites, phosphatidylethanolamine-binding cavity, and the mechanism of enzymatic action. In vitro and in vivo evidence that MCR-5 catalyzes the addition of phosphoethanolamine to the suggestive 4'-phosphate of lipid A moieties is integrated, and results in the consequent polymyxin resistance. In addition, MCR-5 alleviates the colistin-induced formation of reactive oxygen species in E. coli. Taken together, the finding suggests that a growing body of MCR family resistance enzymes are functionally unified.
Keywords: Aeromonas hydrophila; MCR‐5; colistin resistance; functional unification; lipid A; phosphatidylethanolamine (PE) cavity; ping‐pong reaction mechanism; transferable resistance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Ferri M., Ranucci E., Romagnoli P., Giaccone V., Crit. Rev. Food Sci. Nutr. 2017, 57, 2857. - PubMed
-
- WHO , 2014.
-
- Li J., Nation R. L., Turnidge J. D., Milne R. W., Coulthard K., Rayner C. R., Paterson D. L., Lancet Infect. Dis. 2006, 6, 589. - PubMed
-
- Poirel N. K. L., Liassine N., Thanh D., Nordmann P., Lancet Infect. Dis. 2016, 16, 281. - PubMed
LinkOut - more resources
Full Text Sources
