Heterogeneity in surface sensing suggests a division of labor in Pseudomonas aeruginosa populations
- PMID: 31180327
- PMCID: PMC6615863
- DOI: 10.7554/eLife.45084
Heterogeneity in surface sensing suggests a division of labor in Pseudomonas aeruginosa populations
Erratum in
-
Correction: Heterogeneity in surface sensing suggests a division of labor in Pseudomonas aeruginosa populations.Elife. 2020 May 26;9:e59154. doi: 10.7554/eLife.59154. Elife. 2020. PMID: 32452764 Free PMC article.
Abstract
The second messenger signaling molecule cyclic diguanylate monophosphate (c-di-GMP) drives the transition between planktonic and biofilm growth in many bacterial species. Pseudomonas aeruginosa has two surface sensing systems that produce c-di-GMP in response to surface adherence. Current thinking in the field is that once cells attach to a surface, they uniformly respond by producing c-di-GMP. Here, we describe how the Wsp system generates heterogeneity in surface sensing, resulting in two physiologically distinct subpopulations of cells. One subpopulation has elevated c-di-GMP and produces biofilm matrix, serving as the founders of initial microcolonies. The other subpopulation has low c-di-GMP and engages in surface motility, allowing for exploration of the surface. We also show that this heterogeneity strongly correlates to surface behavior for descendent cells. Together, our results suggest that after surface attachment, P. aeruginosa engages in a division of labor that persists across generations, accelerating early biofilm formation and surface exploration.
Keywords: Pseudomonas aeruginosa; Wsp system; biofilm; c-di-gmp; infectious disease; microbiology; surface sensing.
Plain language summary
Bacteria can adopt different lifestyles, depending on the environment in which they grow. They can exist as single cells that are free to explore their environment or group together to form ‘biofilms’. The bacteria in biofilms stick to a surface, and produce a slimy ‘matrix’ that covers and thereby protects them. Biofilms have been found in lung infections that affect people with the genetic disorder cystic fibrosis, and can also form on the surface of medical implants. Because the biofilm lifestyle protects bacteria from the immune system and antimicrobial drugs, learning about how biofilms form could help researchers to discover ways to prevent and treat such infections. Many bacteria switch between the free-living and biofilm lifestyles by altering their levels of a signaling molecule called cyclic diguanylate monophosphate (called c-di-GMP for short). Bacteria living in biofilms have much higher levels of c-di-GMP than their free-living counterparts, and bacteria that have high levels of c-di-GMP produce more biofilm matrix. Bacteria called Pseudomonas aeruginosa use a protein signaling complex called the Wsp system to sense that they are on a surface and increase c-di-GMP production. Questions remained about how quickly this change in production occurs, and whether bacteria pass on their c-di-GMP levels to the new descendant cells when they divide. Armbruster et al. monitored individual cells of P. aeruginosa producing c-di-GMP as they began to form biofilms. Unexpectedly, not all cells increased their c-di-GMP levels when they first attached to a surface. Instead, Armbruster et al. found that there are two populations – high and low c-di-GMP cells – that each perform complementary and important tasks in the early stages of biofilm formation. The high c-di-GMP cells represent ‘biofilm founders’ that start to produce the biofilm matrix, whereas the low c-di-GMP cells represent ‘surface explorers’ that spend more time traveling along the surface. Armbruster et al. found that the Wsp surface sensing system generates these two populations of cells. Moreover, the c-di-GMP levels in a bacterial cell even affect the behavior of the descendant cells that form when it divides. This effect can persist for several cell generations. More work is needed to examine exactly how the biofilm founders and surface explorers interact and influence how biofilms form, and to discover if blocking c-di-GMP signaling prevents biofilm formation. This could ultimately lead to new strategies to prevent and treat infections in humans.
© 2019, Armbruster et al.
Conflict of interest statement
CA, CL, JP, Jd, AX, KZ, BT, LH, FJ, CH, GW, MP No competing interests declared
Figures
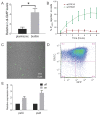
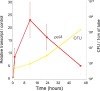

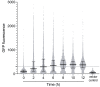
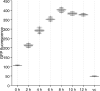
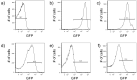
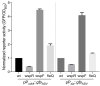
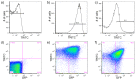
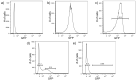
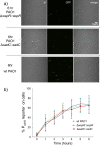
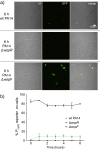
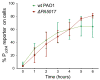
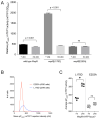



Similar articles
-
Analysis of Pseudomonas aeruginosa c-di-GMP High and Low Subpopulations Using Flow-assisted Cell Sorting (FACS) and Quantitative Reverse Transcriptase PCR (qRT-PCR).Bio Protoc. 2021 Jan 20;11(2):e3891. doi: 10.21769/BioProtoc.3891. eCollection 2021 Jan 20. Bio Protoc. 2021. PMID: 33732780 Free PMC article.
-
Glucose-6-Phosphate Acts as an Extracellular Signal of SagS To Modulate Pseudomonas aeruginosa c-di-GMP Levels, Attachment, and Biofilm Formation.mSphere. 2021 Feb 10;6(1):e01231-20. doi: 10.1128/mSphere.01231-20. mSphere. 2021. PMID: 33568456 Free PMC article.
-
Reduced Intracellular c-di-GMP Content Increases Expression of Quorum Sensing-Regulated Genes in Pseudomonas aeruginosa.Front Cell Infect Microbiol. 2017 Oct 17;7:451. doi: 10.3389/fcimb.2017.00451. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29090193 Free PMC article.
-
Biofilms and Cyclic di-GMP (c-di-GMP) Signaling: Lessons from Pseudomonas aeruginosa and Other Bacteria.J Biol Chem. 2016 Jun 10;291(24):12547-12555. doi: 10.1074/jbc.R115.711507. Epub 2016 Apr 21. J Biol Chem. 2016. PMID: 27129226 Free PMC article. Review.
-
Regulation of biofilm formation in Pseudomonas and Burkholderia species.Environ Microbiol. 2014 Jul;16(7):1961-81. doi: 10.1111/1462-2920.12448. Epub 2014 Mar 31. Environ Microbiol. 2014. PMID: 24592823 Review.
Cited by
-
Phenotypic Patterning through Copy Number Adaptation to Environmental Gradients.ACS Synth Biol. 2024 Mar 15;13(3):728-735. doi: 10.1021/acssynbio.3c00617. Epub 2024 Feb 8. ACS Synth Biol. 2024. PMID: 38330913 Free PMC article.
-
Identification of Cyclic-di-GMP-Modulating Protein Residues by Bidirectionally Evolving a Social Behavior in Pseudomonas fluorescens.mSystems. 2022 Oct 26;7(5):e0073722. doi: 10.1128/msystems.00737-22. Epub 2022 Oct 3. mSystems. 2022. PMID: 36190139 Free PMC article.
-
Single-Molecule Fluorescent In Situ Hybridization (smFISH) for RNA Detection in Bacteria.Methods Mol Biol. 2024;2784:3-23. doi: 10.1007/978-1-0716-3766-1_1. Methods Mol Biol. 2024. PMID: 38502475
-
Diverse Sulfuriferula spp. from sulfide mineral weathering environments oxidize ferrous iron and reduced inorganic sulfur compounds.Appl Environ Microbiol. 2025 Jul 23;91(7):e0021625. doi: 10.1128/aem.00216-25. Epub 2025 Jun 5. Appl Environ Microbiol. 2025. PMID: 40470961 Free PMC article.
-
A motile doublet form of Salmonella Typhimurium diversifies target search behavior at the epithelial surface.Mol Microbiol. 2022 May;117(5):1156-1172. doi: 10.1111/mmi.14898. Epub 2022 Apr 12. Mol Microbiol. 2022. PMID: 35332598 Free PMC article.
References
-
- Armbruster CR, Wolter DJ, Mishra M, Hayden HS, Radey MC, Merrihew G, MacCoss MJ, Burns J, Wozniak DJ, Parsek MR, Hoffman LR. Staphylococcus aureus Protein A Mediates Interspecies Interactions at the Cell Surface of Pseudomonas aeruginosa. mBio. 2016;7 doi: 10.1128/mBio.00538-16. - DOI - PMC - PubMed
-
- Baraquet C, Murakami K, Parsek MR, Harwood CS. The FleQ protein from pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Research. 2012;40:7207–7218. doi: 10.1093/nar/gks384. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K24HL141669/NH/NIH HHS/United States
- K22AI121097/NH/NIH HHS/United States
- GM56665/GM/NIGMS NIH HHS/United States
- T32 GM007270/GM/NIGMS NIH HHS/United States
- R01 AI077628/AI/NIAID NIH HHS/United States
- T32GM007270/NH/NIH HHS/United States
- K24 HL141669/HL/NHLBI NIH HHS/United States
- 1R01AI143730-01/NH/NIH HHS/United States
- R01 AI143916/AI/NIAID NIH HHS/United States
- R01 GM056665/GM/NIGMS NIH HHS/United States
- 5R01AI077628/NH/NIH HHS/United States
- R01AI143916/NH/NIH HHS/United States
- R01 AI134895/AI/NIAID NIH HHS/United States
- K22 AI121097/AI/NIAID NIH HHS/United States
- R01 AI143730/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

