Biogeography of the Oral Microbiome: The Site-Specialist Hypothesis
- PMID: 31180804
- PMCID: PMC7153577
- DOI: 10.1146/annurev-micro-090817-062503
Biogeography of the Oral Microbiome: The Site-Specialist Hypothesis
Abstract
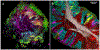
Microbial communities are complex and dynamic, composed of hundreds of taxa interacting across multiple spatial scales. Advances in sequencing and imaging technology have led to great strides in understanding both the composition and the spatial organization of these complex communities. In the human mouth, sequencing results indicate that distinct sites host microbial communities that not only are distinguishable but to a meaningful degree are composed of entirely different microbes. Imaging suggests that the spatial organization of these communities is also distinct. Together, the literature supports the idea that most oral microbes are site specialists. A clear understanding of microbiota structure at different sites in the mouth enables mechanistic studies, informs the generation of hypotheses, and strengthens the position of oral microbiology as a model system for microbial ecology in general.
Keywords: biofilm; fluorescence microscopy; human microbiome; imaging; metagenomics; microbial ecology.
Figures

References
-
- Al-Ahmad A, Follo M, Selzer A-C, Hellwig E, Hannig M, Hannig C. 2009. Bacterial colonization of enamel in situ investigated using fluorescence in situ hybridization. Journal of Medical Microbiology 58: 1359–1366. - PubMed
-
- Al-Ahmad A, Wunder A, Auschill TM, Follo M, Braun G, Hellwig E, Arweiler NB. 2007. The in vivo dynamics of Streptococcus spp., Acinomyces naeslundii, Fusobacterium nucleatum and Veillonella spp. in dental plaque biofilm as analysed by five-colour multiplex fluorescence in situ hybridization. Journal of Medical Microbiology 56: 681–687. - PubMed
-
- Asikainen P, Sirviö E, Mikkonen JJW, Singh SP, Schulten EAJM, ten Bruggenkate CM, Koistinen AP, Kullaa AM. 2015. Microplicae – Specialized Surface Structure of Epithelial Cells of Wet-Surfaced Oral Mucosa. Ultrastructural Pathology 39(5): 299–305. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

