The Ultimate Guide to Bacterial Swarming: An Experimental Model to Study the Evolution of Cooperative Behavior
- PMID: 31180806
- PMCID: PMC7428860
- DOI: 10.1146/annurev-micro-020518-120033
The Ultimate Guide to Bacterial Swarming: An Experimental Model to Study the Evolution of Cooperative Behavior
Abstract
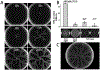
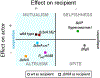
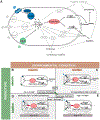
Cooperation has fascinated biologists since Darwin. How did cooperative behaviors evolve despite the fitness cost to the cooperator? Bacteria have cooperative behaviors that make excellent models to take on this age-old problem from both proximate (molecular) and ultimate (evolutionary) angles. We delve into Pseudomonas aeruginosa swarming, a phenomenon where billions of bacteria move cooperatively across distances of centimeters in a matter of a few hours. Experiments with swarming have unveiled a strategy called metabolic prudence that stabilizes cooperation, have showed the importance of spatial structure, and have revealed a regulatory network that integrates environmental stimuli and direct cooperative behavior, similar to a machine learning algorithm. The study of swarming elucidates more than proximate mechanisms: It exposes ultimate mechanisms valid to all scales, from cells in cancerous tumors to animals in large communities.
Keywords: biofilm; cheater; metabolic prudence; rhamnolipids; sociomicrobiology.
Figures
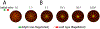


References
-
- Axelrod R and Hamilton WD 1981. The evolution of cooperation. Science 211(4489), pp. 1390–1396. - PubMed
-
- Biddle A, Liang X, Gammon L, Fazil B, Harper LJ, Emich H, Costea DE and Mackenzie IC 2011. Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratory or proliferative. Cancer Research 71(15), pp. 5317–5326. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

