The Microbial Toxin Microcin B17: Prospects for the Development of New Antibacterial Agents
- PMID: 31181289
- PMCID: PMC6722960
- DOI: 10.1016/j.jmb.2019.05.050
The Microbial Toxin Microcin B17: Prospects for the Development of New Antibacterial Agents
Abstract
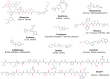
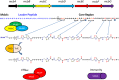
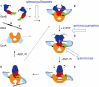
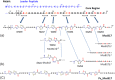
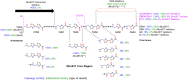
Microcin B17 (MccB17) is an antibacterial peptide produced by strains of Escherichia coli harboring the plasmid-borne mccB17 operon. MccB17 possesses many notable features. It is able to stabilize the transient DNA gyrase-DNA cleavage complex, a very efficient mode of action shared with the highly successful fluoroquinolone drugs. MccB17 stabilizes this complex by a distinct mechanism making it potentially valuable in the fight against bacterial antibiotic resistance. MccB17 was the first compound discovered from the thiazole/oxazole-modified microcins family and the linear azole-containing peptides; these ribosomal peptides are post-translationally modified to convert serine and cysteine residues into oxazole and thiazole rings. These chemical moieties are found in many other bioactive compounds like the vitamin thiamine, the anti-cancer drug bleomycin, the antibacterial sulfathiazole and the antiviral nitazoxanide. Therefore, the biosynthetic machinery that produces these azole rings is noteworthy as a general method to create bioactive compounds. Our knowledge of MccB17 now extends to many aspects of antibacterial-bacteria interactions: production, transport, interaction with its target, and resistance mechanisms; this knowledge has wide potential applicability. After a long time with limited progress on MccB17, recent publications have addressed critical aspects of MccB17 biosynthesis as well as an explosion in the discovery of new related compounds in the thiazole/oxazole-modified microcins/linear azole-containing peptides family. It is therefore timely to summarize the evidence gathered over more than 40 years about this still enigmatic molecule and place it in the wider context of antibacterials.
Keywords: DNA gyrase; DNA topoisomerases; TOMMs; microbial toxins.
Crown Copyright © 2019. Published by Elsevier Ltd. All rights reserved.
Figures





References
-
- Bax R., Green S. Antibiotics: the changing regulatory and pharmaceutical industry paradigm. J. Antimicrob. Chemother. 2015;70:1281–1284. - PubMed
-
- Payne D.J., Gwynn M.N., Holmes D.J., Pompliano D.L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007;6:29–40. - PubMed
-
- Tommasi R., Brown D.G., Walkup G.K., Manchester J.I., Miller A.A. ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discov. 2015;14:529–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/J/00000201/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/P012523/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004561/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J016853/1 /BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical

