Genetic Characterization of a Recombinant Myxoma Virus in the Iberian Hare (Lepus granatensis)
- PMID: 31181645
- PMCID: PMC6631704
- DOI: 10.3390/v11060530
Genetic Characterization of a Recombinant Myxoma Virus in the Iberian Hare (Lepus granatensis)
Abstract
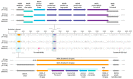
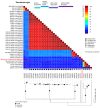
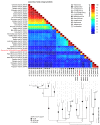
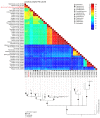
Myxomatosis is a lethal disease in wild European and domestic rabbits (Oryctolagus cuniculus), which is caused by a Myxoma virus (MYXV) infection-a leporipoxvirus that is found naturally in some Sylvilagus rabbit species in South America and California. The introduction of MYXV into feral European rabbit populations of Australia and Europe, in the early 1950s, demonstrated the best-documented field example of host-virus coevolution, following a cross-species transmission. Recently, a new cross-species jump of MYXV has been suggested in both Great Britain and Spain, where European brown hares (Lepus europaeus) and Iberian hares (Lepus granatensis) were found dead with lesions consistent with those observed in myxomatosis. To investigate the possibility of a new cross-species transmission event by MYXV, tissue samples collected from a wild Iberian hare found dead in Spain (Toledo region) were analyzed and deep sequenced. Our results reported a new MYXV isolate (MYXV Toledo) in the tissues of this species. The genome of this new virus was found to encode three disruptive genes (M009L, M036L, and M152R) and a novel ~2.8 kb recombinant region, which resulted from an insertion of four novel poxviral genes towards the 3' end of the negative strand of its genome. From the open reading frames inserted into the MYXV Toledo virus, a new orthologue of a poxvirus host range gene family member was identified, which was related to the MYXV gene M064R. Overall, we confirmed the identity of a new MYXV isolate in Iberian hares, which, we hypothesized, was able to more effectively counteract the host defenses in hares and start an infectious process in this new host.
Keywords: Lepus granatensis; Myxoma virus; poxvirus; recombinant virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Monitoring of emerging myxoma virus epidemics in Iberian hares (Lepus granatensis) in Spain, 2018-2020.Transbound Emerg Dis. 2021 May;68(3):1275-1282. doi: 10.1111/tbed.13781. Epub 2020 Aug 20. Transbound Emerg Dis. 2021. PMID: 32786107
-
Epidemiological surveillance of myxoma virus in European hares (Lepus europaeus) in the Iberian Peninsula: First evidence of infection by the emerging ha-MYXV.Vet Microbiol. 2025 Mar;302:110405. doi: 10.1016/j.vetmic.2025.110405. Epub 2025 Jan 24. Vet Microbiol. 2025. PMID: 39892024
-
Co-infection by classic MYXV and ha-MYXV in Iberian hare (Lepus granatensis) and European wild rabbit (Oryctolagus cuniculus algirus).Transbound Emerg Dis. 2022 Jul;69(4):1684-1690. doi: 10.1111/tbed.14540. Epub 2022 Apr 9. Transbound Emerg Dis. 2022. PMID: 35366052
-
Myxomatosis.Rev Sci Tech. 2015 Aug;34(2):549-56, 539-47. Rev Sci Tech. 2015. PMID: 26601455 Review. English, French.
-
Myxoma virus and the Leporipoxviruses: an evolutionary paradigm.Viruses. 2015 Mar 6;7(3):1020-61. doi: 10.3390/v7031020. Viruses. 2015. PMID: 25757062 Free PMC article. Review.
Cited by
-
Poxvirus cGAMP nucleases: Clues and mysteries from a stolen gene.PLoS Pathog. 2021 Mar 18;17(3):e1009372. doi: 10.1371/journal.ppat.1009372. eCollection 2021 Mar. PLoS Pathog. 2021. PMID: 33735254 Free PMC article. Review. No abstract available.
-
Domestic European Rabbits Oryctolagus cuniculus: A Super-Highway for the Spread of Emergent Viral Diseases to Other Lagomorphs?Transbound Emerg Dis. 2025 Jun 6;2025:1129135. doi: 10.1155/tbed/1129135. eCollection 2025. Transbound Emerg Dis. 2025. PMID: 40521303 Free PMC article.
-
Identification of a Novel Myxoma Virus C7-Like Host Range Factor That Enabled a Species Leap from Rabbits to Hares.mBio. 2022 Apr 26;13(2):e0346121. doi: 10.1128/mbio.03461-21. Epub 2022 Mar 30. mBio. 2022. PMID: 35352978 Free PMC article.
-
Retrospective serological and molecular survey of myxoma or antigenically related virus in the Iberian hare, Lepus granatensis.Transbound Emerg Dis. 2022 Nov;69(6):3637-3650. doi: 10.1111/tbed.14734. Epub 2022 Oct 26. Transbound Emerg Dis. 2022. PMID: 36219552 Free PMC article.
-
Identification and Characterisation of a Myxoma Virus Detected in the Italian Hare (Lepus corsicanus).Viruses. 2024 Mar 12;16(3):437. doi: 10.3390/v16030437. Viruses. 2024. PMID: 38543802 Free PMC article.
References
-
- Fenner F., Ratcliffe F.N. Myxomatosis. Cambridge University Press; Cambridge, UK: 1965.
-
- Wilkinson L. Biological control of vertebrate pests: The history of myxomatosis, an experiment in evolution. Med. Hist. 2001;45:139–140. doi: 10.1017/S0025727300067594. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

