Emerging Novel GII.P16 Noroviruses Associated with Multiple Capsid Genotypes
- PMID: 31181749
- PMCID: PMC6631344
- DOI: 10.3390/v11060535
Emerging Novel GII.P16 Noroviruses Associated with Multiple Capsid Genotypes
Abstract
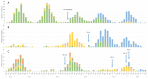
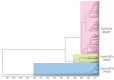
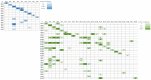
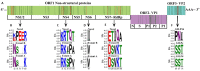
Noroviruses evolve by antigenic drift and recombination, which occurs most frequently at the junction between the non-structural and structural protein coding genomic regions. In 2015, a novel GII.P16-GII.4 Sydney recombinant strain emerged, replacing the predominance of GII.Pe-GII.4 Sydney among US outbreaks. Distinct from GII.P16 polymerases detected since 2010, this novel GII.P16 was subsequently detected among GII.1, GII.2, GII.3, GII.10 and GII.12 viruses, prompting an investigation on the unique characteristics of these viruses. Norovirus positive samples (n = 1807) were dual-typed, of which a subset (n = 124) was sequenced to yield near-complete genomes. CaliciNet and National Outbreak Reporting System (NORS) records were matched to link outbreak characteristics and case outcomes to molecular data and GenBank was mined for contextualization. Recombination with the novel GII.P16 polymerase extended GII.4 Sydney predominance and increased the number of GII.2 outbreaks in the US. Introduction of the novel GII.P16 noroviruses occurred without unique amino acid changes in VP1, more severe case outcomes, or differences in affected population. However, unique changes were found among NS1/2, NS4 and VP2 proteins, which have immune antagonistic functions, and the RdRp. Multiple polymerase-capsid combinations were detected among GII viruses including 11 involving GII.P16. Molecular surveillance of protein sequences from norovirus genomes can inform the functional importance of amino acid changes in emerging recombinant viruses and aid in vaccine and antiviral formulation.
Keywords: GII.4 Sydney; GII.P16; Norovirus; clinical outcomes; dual-typing; herd immunity; immune antagonism; molecular epidemiology; non-structural proteins; recombinants.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Genetic and Epidemiologic Trends of Norovirus Outbreaks in the United States from 2013 to 2016 Demonstrated Emergence of Novel GII.4 Recombinant Viruses.J Clin Microbiol. 2017 Jul;55(7):2208-2221. doi: 10.1128/JCM.00455-17. Epub 2017 May 10. J Clin Microbiol. 2017. PMID: 28490488 Free PMC article.
-
Characterization of the complete genome sequence of the recombinant norovirus GII.P16/GII.4_Sydney_2012 revealed in Russia.Vavilovskii Zhurnal Genet Selektsii. 2020 Feb;24(1):69-79. doi: 10.18699/VJ20.597. Vavilovskii Zhurnal Genet Selektsii. 2020. PMID: 33659783 Free PMC article.
-
Emergence of novel recombinant GII.P16_GII.2 and GII. P16_GII.4 Sydney 2012 norovirus strains in Italy, winter 2016/2017.New Microbiol. 2018 Jan;41(1):71-72. New Microbiol. 2018. PMID: 29505067
-
Global and regional circulation trends of norovirus genotypes and recombinants, 1995-2019: A comprehensive review of sequences from public databases.Rev Med Virol. 2022 Sep;32(5):e2354. doi: 10.1002/rmv.2354. Epub 2022 Apr 28. Rev Med Virol. 2022. PMID: 35481689 Free PMC article. Review.
-
Emergence of norovirus strains: A tale of two genes.Virus Evol. 2019 Nov 25;5(2):vez048. doi: 10.1093/ve/vez048. eCollection 2019 Jul. Virus Evol. 2019. PMID: 32161666 Free PMC article. Review.
Cited by
-
A new wave of resurgence for GII.4 Sydney in Huzhou, particularly GII.4 Sydney[P16], between 2019 and 2023.BMC Infect Dis. 2025 Feb 19;25(1):241. doi: 10.1186/s12879-025-10648-5. BMC Infect Dis. 2025. PMID: 39972255 Free PMC article.
-
Minimal Antigenic Evolution after a Decade of Norovirus GII.4 Sydney_2012 Circulation in Humans.J Virol. 2023 Feb 28;97(2):e0171622. doi: 10.1128/jvi.01716-22. Epub 2023 Jan 23. J Virol. 2023. PMID: 36688654 Free PMC article.
-
Emergence of GII.4 Sydney[P16]-like Norovirus-Associated Gastroenteritis, China, 2020-2022.Emerg Infect Dis. 2023 Sep;29(9):1837-1841. doi: 10.3201/eid2909.230383. Emerg Infect Dis. 2023. PMID: 37610173 Free PMC article.
-
Simvastatin Reduces Protection and Intestinal T Cell Responses Induced by a Norovirus P Particle Vaccine in Gnotobiotic Pigs.Pathogens. 2021 Jul 1;10(7):829. doi: 10.3390/pathogens10070829. Pathogens. 2021. PMID: 34357979 Free PMC article.
-
Epidemiological and molecular surveillance of norovirus in the Brazilian Amazon: description of recombinant genotypes and improvement of evolutionary analysis.Rev Inst Med Trop Sao Paulo. 2024 Apr 19;66:e22. doi: 10.1590/S1678-9946202466022. eCollection 2024. Rev Inst Med Trop Sao Paulo. 2024. PMID: 38656038 Free PMC article.
References
-
- Kirk M.D., Pires S.M., Black R.E., Caipo M., Crump J.A., Devleesschauwer B., Dopfer D., Fazil A., Fischer-Walker C.L., Hald T., et al. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLoS Med. 2015;12:e1001921. - PMC - PubMed
-
- Wikswo M.E., Kambhampati A., Shioda K., Walsh K.A., Bowen A., Hall A.J. Outbreaks of Acute Gastroenteritis Transmitted by Person-to-Person Contact, Environmental Contamination, and Unknown Modes of Transmission—United States, 2009–2013. Morb. Mortal. Wkly. Rep. Surveill. Summ. 2015;64:1–16. doi: 10.15585/mmwr.ss6412a1. - DOI - PubMed
-
- Shah M.P., Wikswo M.E., Barclay L., Kambhampati A., Shioda K., Parashar U.D., Vinje J., Hall A.J. Near Real-Time Surveillance of U.S. Norovirus Outbreaks by the Norovirus Sentinel Testing and Tracking Network—United States, August 2009–July 2015. MMWR Morb. Mortal. Wkly. Rep. 2017;66:185–189. doi: 10.15585/mmwr.mm6607a1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

